1Y94
 
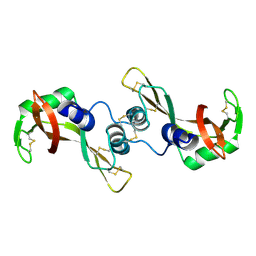 | | Crystal structure of the G16S/N17T/P19A/S20A/N67D Variant Of Bovine seminal Ribonuclease | | Descriptor: | Seminal ribonuclease | | Authors: | Picone, D, Di Fiore, A, Ercole, C, Franzese, M, Sica, F, Tomaselli, S, Mazzarella, L. | | Deposit date: | 2004-12-14 | | Release date: | 2004-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Role of the Hinge Loop in Domain Swapping: THE SPECIAL CASE OF BOVINE SEMINAL RIBONUCLEASE.
J.Biol.Chem., 280, 2005
|
|
3H98
 
 | | Crystal structure of HCV NS5b 1b with (1,1-dioxo-2H-[1,2,4]benzothiadiazin-3-yl) azolo[1,5-a]pyrimidine derivative | | Descriptor: | GLYCEROL, N-{3-[5-hydroxy-8-(3-methylbutyl)-7-oxo-7,8-dihydroimidazo[1,2-a]pyrimidin-6-yl]-1,1-dioxido-4H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Wang, G, Lei, H, Wang, X, Das, D, Mackinnon, C, Montalbetti, C.A.G, Mears, R, Gai, X, Bailey, S, Ruhrmund, D, Hooi, L, Misialek, S, Rajagopalan, R, Cheng, R.K.Y, Barker, J.L, Felicetti, B, Stoycheva, A, Buckman, B, Kossen, K, Seiwert, S, Beigelmana, L. | | Deposit date: | 2009-04-30 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | HCV NS5B polymerase inhibitors 2: Synthesis and in vitro activity of (1,1-dioxo-2H-[1,2,4]benzothiadiazin-3-yl) azolo[1,5-a]pyridine and azolo[1,5-a]pyrimidine derivatives.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1FYD
 
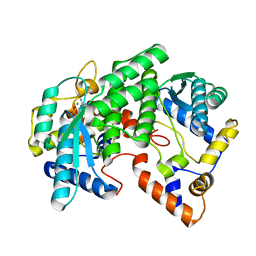 | | CRYSTAL STRUCTURE OF NH3-DEPENDENT NAD+ SYNTHETASE FROM BACILLUS SUBTILIS COMPLEXED WITH ONE MOLECULE AMP, ONE PYROPHOSPHATE ION AND ONE MG2+ ION | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, NH(3)-DEPENDENT NAD(+) SYNTHETASE, ... | | Authors: | Devedjiev, Y, Symersky, J, Singh, R, Brouillette, W, Muccio, D, Jedrzejas, M, Brouillette, C, DeLucas, L. | | Deposit date: | 2000-09-28 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Stabilization of active-site loops in NH3-dependent NAD+ synthetase from Bacillus subtilis.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
4CSS
 
 | | Crystal structure of FimH in complex with a sulfonamide biphenyl alpha D-mannoside | | Descriptor: | 4'-(alpha-D-Mannopyranosyloxy)-biphenyl-4-methyl sulfonamide, GLYCEROL, PROTEIN FIMH | | Authors: | Kleeb, S, Pang, L, Mayer, K, Sigl, A, Eris, D, Preston, R.C, Zihlmann, P, Abgottspon, D, Hutter, A, Scharenberg, M, Jian, X, Navarra, G, Rabbani, S, Smiesko, M, Luedin, N, Jakob, R.P, Schwardt, O, Maier, T, Sharpe, T, Ernst, B. | | Deposit date: | 2014-03-10 | | Release date: | 2015-02-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.069 Å) | | Cite: | Fimh Antagonists: Bioisosteres to Improve the in Vitro and in Vivo Pk/Pd Profile.
J.Med.Chem., 58, 2015
|
|
4CHC
 
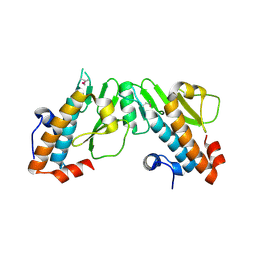 | | Crystal structure of the N-terminal domain of the PA subunit of Thogoto virus polymerase (form 2) | | Descriptor: | POLYMERASE ACIDIC PROTEIN | | Authors: | Guilligay, D, Kadlec, J, Crepin, T, Lunardi, T, Bouvier, D, Kochs, G, Ruigrok, R.W.H, Cusack, S. | | Deposit date: | 2013-12-01 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Comparative Structural and Functional Analysis of Orthomyxovirus Polymerase CAP-Snatching Domains.
Plos One, 9, 2014
|
|
4RED
 
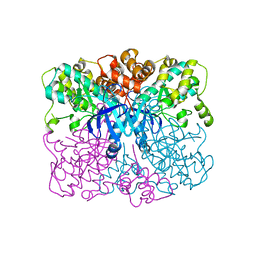 | | Crystal structure of human AMPK alpha1 KD-AID with K43A mutation | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1 | | Authors: | Zhou, X.E, Ke, J, Li, X, Wang, L, Gu, X, de Waal, P.W, Tan, M.H.E, Wang, D, Wu, D, Xu, H.E, Melcher, K. | | Deposit date: | 2014-09-22 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis of AMPK regulation by adenine nucleotides and glycogen.
Cell Res., 25, 2015
|
|
7F3Y
 
 | | Wild-type Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with methotrexate (MTX), NADPH and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, GLYCEROL, ... | | Authors: | Vanichtanankul, J, Tanramluk, D, Yuvaniyama, J, Yuthavong, Y. | | Deposit date: | 2021-06-17 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | MANORAA: A machine learning platform to guide protein-ligand design by anchors and influential distances.
Structure, 30, 2022
|
|
7F3Z
 
 | | Double mutant Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS-K1, C59R+S108N) complexed with Trimethoprim (TOP), NADPH and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, GLYCEROL, ... | | Authors: | Vanichtanankul, J, Tanramluk, D, Chitnumsub, P, Yuvaniyama, J, Yuthavong, Y. | | Deposit date: | 2021-06-17 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | MANORAA: A machine learning platform to guide protein-ligand design by anchors and influential distances.
Structure, 30, 2022
|
|
2CSC
 
 | |
4CHF
 
 | | Crystal structure of the putative cap-binding domain of the PB2 subunit of Thogoto virus polymerase (form 2) | | Descriptor: | POLYMERASE BASIC PROTEIN 2 | | Authors: | Guilligay, D, Kadlec, J, Crepin, T, Lunardi, T, Bouvier, D, Kochs, G, Ruigrok, R.W.H, Cusack, S. | | Deposit date: | 2013-12-01 | | Release date: | 2014-02-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Comparative Structural and Functional Analysis of Orthomyxovirus Polymerase CAP-Snatching Domains.
Plos One, 9, 2014
|
|
1DBR
 
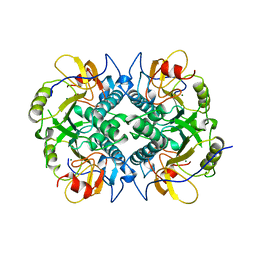 | | HYPOXANTHINE GUANINE XANTHINE | | Descriptor: | HYPOXANTHINE GUANINE XANTHINE PHOSPHORIBOSYLTRANSFERASE, MAGNESIUM ION | | Authors: | Schumacher, M.A, Carter, D, Roos, D, Ullman, B, Brennan, R.G. | | Deposit date: | 1996-02-13 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Toxoplasma gondii HGXPRTase reveal the catalytic role of a long flexible loop.
Nat.Struct.Biol., 3, 1996
|
|
4CDL
 
 | | Crystal Structure of Retro-aldolase RA110.4-6 Complexed with Inhibitor 1-(6-methoxy-2-naphthalenyl)-1,3-butanedione | | Descriptor: | (2E)-1-(6-methoxynaphthalen-2-yl)but-2-en-1-one, STEROID DELTA-ISOMERASE | | Authors: | Pinkas, D.M, Studer, S, Obexer, R, Giger, L, Gruetter, M.G, Baker, D, Hilvert, D. | | Deposit date: | 2013-11-01 | | Release date: | 2014-11-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Active Site Plasticity of a Computationally Designed Retro-Aldolase Enzyme
To be Published
|
|
4CFF
 
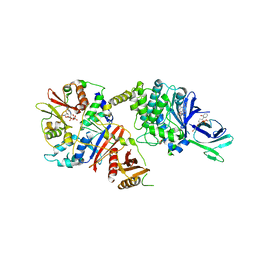 | | Structure of full length human AMPK in complex with a small molecule activator, a thienopyridone derivative (A-769662) | | Descriptor: | 3-[4-(2-hydroxyphenyl)phenyl]-4-oxidanyl-6-oxidanylidene-7H-thieno[2,3-b]pyridine-5-carbonitrile, 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-1, ... | | Authors: | Xiao, B, Sanders, M.J, Carmena, D, Bright, N.J, Haire, L.F, Underwood, E, Patel, B.R, Heath, R.B, Walker, P.A, Hallen, S, Giordanetto, F, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2013-11-14 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.924 Å) | | Cite: | Structural Basis of Ampk Regulation by Small Molecule Activators.
Nat.Commun., 4, 2013
|
|
7FDS
 
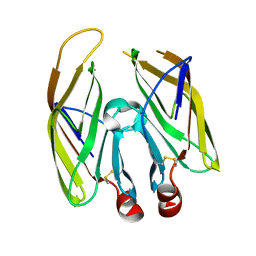 | | High resolution crystal structure of LpqH from Mycobacterium tuberculosis | | Descriptor: | Lipoprotein LpqH | | Authors: | Kundapura, S.V, Chatterjee, S, Samanta, D, Ramagopal, U.A. | | Deposit date: | 2021-07-17 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.258 Å) | | Cite: | High-resolution crystal structure of LpqH, an immunomodulatory surface lipoprotein of Mycobacterium tuberculosis reveals a distinct fold and a conserved cleft on its surface.
Int.J.Biol.Macromol., 210, 2022
|
|
4CST
 
 | | Crystal structure of FimH in complex with 3'-Chloro-4'-(alpha-D-mannopyranosyloxy)-biphenyl-4-carbonitrile | | Descriptor: | 3'-chloro-4'-(alpha-D-mannopyranosyloxy)biphenyl-4-carbonitrile, PROTEIN FIMH | | Authors: | Kleeb, S, Pang, L, Mayer, K, Sigl, A, Eris, D, Preston, R.C, Zihlmann, P, Abgottspon, D, Hutter, A, Scharenberg, M, Jian, X, Navarra, G, Rabbani, S, Smiesko, M, Luedin, N, Jakob, R.P, Schwardt, O, Maier, T, Sharpe, T, Ernst, B. | | Deposit date: | 2014-03-10 | | Release date: | 2015-02-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Fimh Antagonists: Bioisosteres to Improve the in Vitro and in Vivo Pk/Pd Profile.
J.Med.Chem., 58, 2015
|
|
1XYC
 
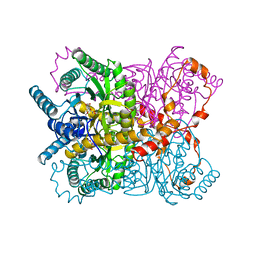 | | X-RAY CRYSTALLOGRAPHIC STRUCTURES OF D-XYLOSE ISOMERASE-SUBSTRATE COMPLEXES POSITION THE SUBSTRATE AND PROVIDE EVIDENCE FOR METAL MOVEMENT DURING CATALYSIS | | Descriptor: | 3-O-METHYLFRUCTOSE IN LINEAR FORM, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Lavie, A, Allen, K.N, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-01-03 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | X-ray crystallographic structures of D-xylose isomerase-substrate complexes position the substrate and provide evidence for metal movement during catalysis.
Biochemistry, 33, 1994
|
|
4CHD
 
 | | Crystal structure of the '627' domain of the PB2 subunit of Thogoto virus polymerase | | Descriptor: | POLYMERASE ACIDIC PROTEIN | | Authors: | Guilligay, D, Kadlec, J, Crepin, T, Lunardi, T, Bouvier, D, Kochs, G, Ruigrok, R.W.H, Cusack, S. | | Deposit date: | 2013-12-01 | | Release date: | 2014-02-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Comparative Structural and Functional Analysis of Orthomyxovirus Polymerase CAP-Snatching Domains.
Plos One, 9, 2014
|
|
1XYL
 
 | | THE ROLE OF THE DIVALENT METAL ION IN SUGAR BINDING, RING OPENING, AND ISOMERIZATION BY D-XYLOSE ISOMERASE: REPLACEMENT OF A CATALYTIC METAL BY AN AMINO-ACID | | Descriptor: | HYDROXIDE ION, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Allen, K.N, Lavie, A, Petsko, G.A, Ringe, D. | | Deposit date: | 1993-12-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of the divalent metal ion in sugar binding, ring opening, and isomerization by D-xylose isomerase: replacement of a catalytic metal by an amino acid.
Biochemistry, 33, 1994
|
|
2HFD
 
 | | NMR structure of protein Hydrogenase-1 operon protein hyaE from Escherichia coli: Northeast Structural Genomics Consortium Target ER415 | | Descriptor: | Hydrogenase-1 operon protein hyaE | | Authors: | Singarapu, K.K, Liu, G, Eletsky, A, Parish, D, Atreya, H.S, Xu, D, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-23 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Protein chaperones Q8ZP25_SALTY from Salmonella typhimurium and HYAE_ECOLI from Escherichia coli exhibit thioredoxin-like structures despite lack of canonical thioredoxin active site sequence motif.
J.STRUCT.FUNCT.GENOM., 9, 2008
|
|
2MWS
 
 | |
1YQ4
 
 | | Avian respiratory complex ii with 3-nitropropionate and ubiquinone | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 3-NITROPROPANOIC ACID, Coenzyme Q10, ... | | Authors: | Huang, L, Sun, G, Cobessi, D, Wang, A, Shen, J.T, Tung, E.Y, Anderson, V.E, Berry, E.A. | | Deposit date: | 2005-02-01 | | Release date: | 2005-12-20 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | 3-Nitropropionic Acid Is a Suicide Inhibitor of Mitochondrial Respiration That, upon Oxidation by Complex II, Forms a Covalent Adduct with a Catalytic Base Arginine in the Active Site of the Enzyme
J.Biol.Chem., 281, 2006
|
|
1FCY
 
 | | ISOTYPE SELECTIVITY OF THE HUMAN RETINOIC ACID NUCLEAR RECEPTOR HRAR: THE COMPLEX WITH THE RARBETA/GAMMA-SELECTIVE RETINOID CD564 | | Descriptor: | 6-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTALENE-2-CARBONYL)-NAPHTALENE-2-CARBOXYLIC ACID, DODECYL-ALPHA-D-MALTOSIDE, RETINOIC ACID RECEPTOR GAMMA-1 | | Authors: | Klaholz, B.P, Mitschler, A, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-07-19 | | Release date: | 2000-09-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for isotype selectivity of the human retinoic acid nuclear receptor.
J.Mol.Biol., 302, 2000
|
|
1GH1
 
 | | NMR STRUCTURES OF WHEAT NONSPECIFIC LIPID TRANSFER PROTEIN | | Descriptor: | NONSPECIFIC LIPID TRANSFER PROTEIN | | Authors: | Gincel, E, Simorre, J.P, Caille, A, Marion, D, Ptak, M, Vovelle, F. | | Deposit date: | 2000-10-29 | | Release date: | 2000-11-22 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure in solution of a wheat lipid-transfer protein from multidimensional 1H-NMR data. A new folding for lipid carriers.
Eur.J.Biochem., 226, 1994
|
|
2CBX
 
 | | X-ray crystal structure of 5'-fluorodeoxyadenosine synthase from Streptomyces cattleya complexed with beta-D-erythrofuranosyl- adenosine | | Descriptor: | 5'-FLUORO-5'-DEOXYADENOSINE SYNTHASE, BETA-D-ERYTHROFURANOSYL-ADENOSINE, GLYCEROL | | Authors: | McEwan, A.R, Cadicamo, C.D, Deng, H, McGlinchey, R.P, Robinson, D.R, O'Hagan, D, Naismith, J.H, Spencer, J. | | Deposit date: | 2006-01-09 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate specificity in enzymatic fluorination. The fluorinase from Streptomyces cattleya accepts 2'-deoxyadenosine substrates.
Org. Biomol. Chem., 4, 2006
|
|
2L31
 
 | | Human PARP-1 zinc finger 2 | | Descriptor: | Poly [ADP-ribose] polymerase 1, ZINC ION | | Authors: | Neuhaus, D, Eustermann, S, Yang, J, Videler, H. | | Deposit date: | 2010-08-30 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The DNA-binding domain of human PARP-1 interacts with DNA single-strand breaks as a monomer through its second zinc finger.
J.Mol.Biol., 407, 2011
|
|
