3EMN
 
 | | The Crystal Structure of Mouse VDAC1 at 2.3 A resolution | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Voltage-dependent anion-selective channel protein 1 | | Authors: | Ujwal, R, Cascio, D, Colletier, J.-P, Faham, S, Zhang, J, Toro, L, Ping, P, Abramson, J. | | Deposit date: | 2008-09-24 | | Release date: | 2008-12-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of mouse VDAC1 at 2.3 A resolution reveals mechanistic insights into metabolite gating
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2ESY
 
 | | Structure and influence on stability and activity of the N-terminal propetide part of lung surfactant protein C | | Descriptor: | lung surfactant protein C | | Authors: | Li, J, Liepinsh, E, Almlen, A, Thyberg, J, Curstedt, T, Jornvall, H, Johansson, J. | | Deposit date: | 2005-10-27 | | Release date: | 2005-11-15 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structure and influence on stability and activity of the N-terminal propeptide part of lung surfactant protein C
Febs J., 273, 2006
|
|
3GIN
 
 | |
1M56
 
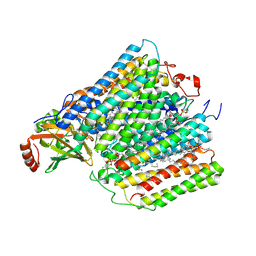 | | Structure of cytochrome c oxidase from Rhodobactor sphaeroides (Wild Type) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Svensson-Ek, M, Abramson, J, Larsson, G, Tornroth, S, Brezezinski, P, Iwata, S. | | Deposit date: | 2002-07-08 | | Release date: | 2002-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The X-ray crystal structures of wild-type and EQ(I-286) mutant cytochrome c oxidases from Rhodobacter sphaeroides.
J.Mol.Biol., 321, 2002
|
|
3P59
 
 | | First Crystal Structure of a RNA Nanosquare | | Descriptor: | COBALT HEXAMMINE(III), MAGNESIUM ION, RNA (5'-R(*CP*CP*GP*GP*AP*GP*GP*AP*AP*CP*UP*AP*CP*(5BU)P*G)-3'), ... | | Authors: | Dibrov, S, Hermann, T, McLean, J, Parsons, J. | | Deposit date: | 2010-10-08 | | Release date: | 2011-04-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1793 Å) | | Cite: | Self-assembling RNA square.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3RYO
 
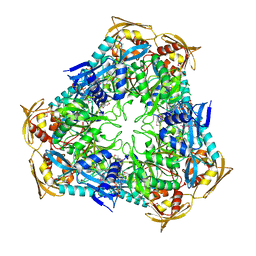 | | Crystal Structure of Enhanced Intracellular Survival (Eis) Protein from Mycobacterium tuberculosis with Acetyl CoA | | Descriptor: | ACETYL COENZYME *A, Enhanced intracellular survival protein | | Authors: | Kim, K.H, An, D.R, Yoon, J.Y, Kim, H.S, Yoon, H.J, Song, J, Im, H.N, Kim, J, Kim, D.J, Lee, S.J, Kim, H.J, Lee, J.Y, Suh, S.W. | | Deposit date: | 2011-05-11 | | Release date: | 2012-05-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mycobacterium tuberculosis Eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7
Proc.Natl.Acad.Sci.USA, 2012
|
|
1M57
 
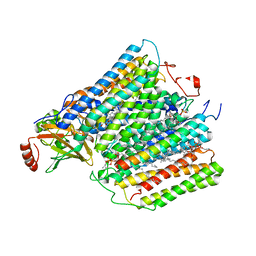 | | Structure of cytochrome c oxidase from Rhodobacter sphaeroides (EQ(I-286) mutant)) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Svensson-Ek, M, Abramson, J, Larsson, G, Tornroth, S, Brezezinski, P, Iwata, S. | | Deposit date: | 2002-07-08 | | Release date: | 2002-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The X-ray crystal structures of wild-type and EQ(I-286) mutant cytochrome c oxidases from Rhodobacter sphaeroides.
J.Mol.Biol., 321, 2002
|
|
4IPN
 
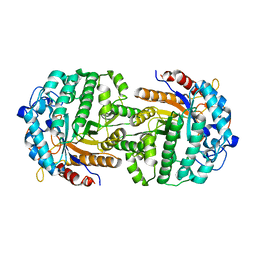 | | The complex structure of 6-phospho-beta-glucosidase BglA-2 with thiocellobiose-6P from Streptococcus pneumoniae | | Descriptor: | 6-O-phosphono-alpha-L-idopyranose-(1-4)-4-thio-beta-D-glucopyranose, 6-phospho-beta-glucosidase | | Authors: | Yu, W.L, Jiang, Y.L, Andreas, P, Cheng, W, Bai, X.H, Ren, Y.M, Thompsonn, J, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2013-01-10 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.411 Å) | | Cite: | Structural insights into the substrate specificity of a 6-phospho-&[beta]-glucosidase BglA-2 from Streptococcus pneumoniae TIGR4
J.Biol.Chem., 288, 2013
|
|
3GL6
 
 | |
1TRJ
 
 | | Homology Model of Yeast RACK1 Protein fitted into 11.7A cryo-EM map of Yeast 80S Ribosome | | Descriptor: | Guanine nucleotide-binding protein beta subunit-like protein, Helix 39 of 18S rRNA, Helix 40 of 18S rRNA | | Authors: | Sengupta, J, Nilsson, J, Gursky, R, Spahn, C.M, Nissen, P, Frank, J. | | Deposit date: | 2004-06-21 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM
Nat.Struct.Mol.Biol., 11, 2004
|
|
1XDG
 
 | | X-ray structure of LFA-1 I-domain in complex with LFA878 at 2.1A resolution | | Descriptor: | (1S,3R,8AS)-8-(2-{(4S,6S)-3-(4-HYDROXY-3-METHOXYBENZYL)-4-[2-(METHYLAMINO)-2-OXOETHYL]-2-OXO-1,3-OXAZINAN-6-YL}ETHYL)-3 ,7-DIMETHYL-1,2,3,7,8,8A-HEXAHYDRONAPHTHALEN-1-YL (2R)-2-METHYLBUTANOATE, Integrin alpha-L, MAGNESIUM ION | | Authors: | Weitz-Schmidt, G, Welzenbach, K, Dawson, J, Kallen, J. | | Deposit date: | 2004-09-06 | | Release date: | 2004-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Improved lymphocyte function-associated antigen-1 (LFA-1) inhibition by statin derivatives: molecular basis determined by x-ray analysis and monitoring of LFA-1 conformational changes in vitro and ex vivo
J.Biol.Chem., 279, 2004
|
|
4K44
 
 | |
2F74
 
 | | Murine MHC class I H-2Db in complex with human b2-microglobulin and LCMV-derived immunodminant peptide gp33 | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Achour, A, Michaelsson, J, Harris, R.A, Ljunggren, H.G, Karre, K, Schneider, G, Sandalova, T. | | Deposit date: | 2005-11-30 | | Release date: | 2006-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of the Differential Stability and Receptor Specificity of H-2D(b) in Complex with Murine versus Human beta(2)-Microglobulin.
J.Mol.Biol., 356, 2006
|
|
1XDD
 
 | | X-ray structure of LFA-1 I-domain in complex with LFA703 at 2.2A resolution | | Descriptor: | 8-[2-((2S)-4-HYDROXY-1-{[5-(HYDROXYMETHYL)-6-METHOXY-2-NAPHTHYL]METHYL}-6-OXOPIPERIDIN-2-YL)ETHYL]-3,7-DIMETHYL-1,2,3,7,8,8A-HEXAHYDRONAPHTHALEN-1-YL 2-METHYLBUTANOATE, Integrin alpha-L, MAGNESIUM ION | | Authors: | Weitz-Schmidt, G, Welzenbach, K, Dawson, J, Kallen, J. | | Deposit date: | 2004-09-06 | | Release date: | 2004-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Improved lymphocyte function-associated antigen-1 (LFA-1) inhibition by statin derivatives: molecular basis determined by x-ray analysis and monitoring of LFA-1 conformational changes in vitro and ex vivo
J.Biol.Chem., 279, 2004
|
|
4IPL
 
 | | The crystal structure of 6-phospho-beta-glucosidase BglA-2 from Streptococcus pneumoniae | | Descriptor: | 6-phospho-beta-glucosidase, GLYCEROL | | Authors: | Yu, W.L, Jiang, Y.L, Andreas, P, Cheng, W, Bai, X.H, Ren, Y.M, Thompsonn, J, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2013-01-10 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Structural insights into the substrate specificity of a 6-phospho-&[beta]-glucosidase BglA-2 from Streptococcus pneumoniae TIGR4
J.Biol.Chem., 288, 2013
|
|
3RFH
 
 | |
4L67
 
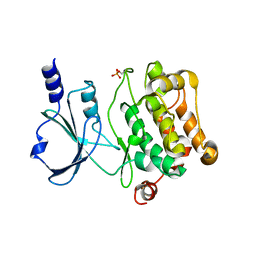 | | Crystal Structure of Catalytic Domain of PAK4 | | Descriptor: | Serine/threonine-protein kinase PAK 4 | | Authors: | Wang, W, Song, J. | | Deposit date: | 2013-06-12 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | NMR binding and crystal structure reveal that intrinsically-unstructured regulatory domain auto-inhibits PAK4 by a mechanism different for that of PAK1
Biochem.Biophys.Res.Commun., 438, 2013
|
|
3JD0
 
 | | Glutamate dehydrogenase in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JD4
 
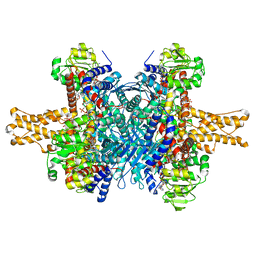 | | Glutamate dehydrogenase in complex with NADH and GTP, closed conformation | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, ... | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JD1
 
 | | Glutamate dehydrogenase in complex with NADH, closed conformation | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3RFG
 
 | |
3JCZ
 
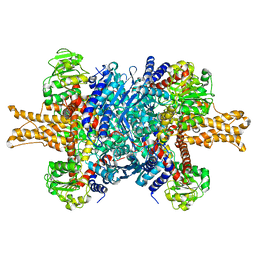 | | Structure of bovine glutamate dehydrogenase in the unliganded state | | Descriptor: | Glutamate dehydrogenase 1, mitochondrial | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-27 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JD3
 
 | | Glutamate dehydrogenase in complex with NADH and GTP, open conformation | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, ... | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
7CJW
 
 | | Solution structure of monomeric superoxide dismutase 1 with an additional mutation H46W in a crowded environment | | Descriptor: | Monomeric Human Cu,Zn Superoxide dismutase | | Authors: | Iwakawa, N, Morimoto, D, Walinda, E, Danielsson, J, Shirakawa, M, Sugase, K. | | Deposit date: | 2020-07-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Transient Diffusive Interactions with a Protein Crowder Affect Aggregation Processes of Superoxide Dismutase 1 beta-Barrel.
J.Phys.Chem.B, 125, 2021
|
|
3OPE
 
 | | Structural Basis of Auto-inhibitory mechanism of Histone methyltransferase | | Descriptor: | Probable histone-lysine N-methyltransferase ASH1L, S-ADENOSYLMETHIONINE, ZINC ION | | Authors: | An, S, Song, J. | | Deposit date: | 2010-08-31 | | Release date: | 2011-01-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the human histone methyltransferase ASH1L catalytic domain and its implications for the regulatory mechanism
J.Biol.Chem., 286, 2011
|
|
