7DRR
 
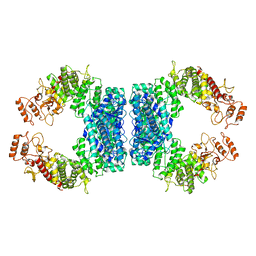 | | Structure of SspE-R100A protein | | Descriptor: | SspE protein | | Authors: | Haiyan, G, Jinchuan, Z, Chen, S, Wang, L, Wu, G. | | Deposit date: | 2020-12-29 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Nicking mechanism underlying the DNA phosphorothioate-sensing antiphage defense by SspE.
Nat Commun, 13, 2022
|
|
7DRS
 
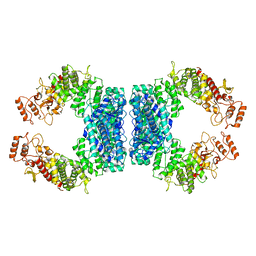 | | Structure of SspE_40224 | | Descriptor: | SspE protein | | Authors: | Haiyan, G, Jinchuan, Z, Chen, S, Wang, L, Wu, G. | | Deposit date: | 2020-12-29 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Nicking mechanism underlying the DNA phosphorothioate-sensing antiphage defense by SspE.
Nat Commun, 13, 2022
|
|
7E5S
 
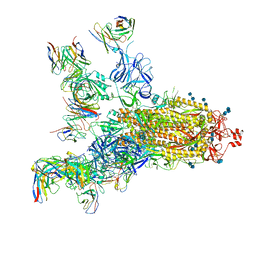 | | SARS-CoV-2 S trimer with four-antibody cocktail complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FC05 heavy chain, FC05 light chain, ... | | Authors: | Sun, Y, Wang, L, Wang, N, Feng, R, Wang, X. | | Deposit date: | 2021-02-20 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure-based development of three- and four-antibody cocktails against SARS-CoV-2 via multiple mechanisms.
Cell Res., 31, 2021
|
|
9ASB
 
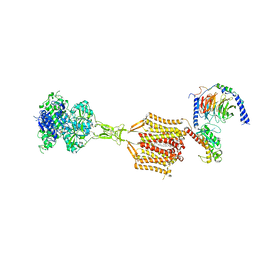 | | Structure of human calcium-sensing receptor in complex with chimeric Gq (miniGisq) protein in nanodiscs | | Descriptor: | (19R,22S,25R)-22,25,26-trihydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphahexacosan-19-yl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zuo, H, Park, J, Frangaj, A, Ye, J, Lu, G, Manning, J.J, Asher, W.B, Lu, Z, Hu, G, Wang, L, Mendez, J, Eng, E, Zhang, Z, Lin, X, Grasucci, R, Hendrickson, W.A, Clarke, O.B, Javitch, J.A, Conigrave, A.D, Fan, Q.R. | | Deposit date: | 2024-02-24 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Promiscuous G-protein activation by the calcium-sensing receptor.
Nature, 629, 2024
|
|
9AVG
 
 | | Structure of human calcium-sensing receptor in complex with chimeric Gs (miniGis) protein in nanodiscs | | Descriptor: | (19R,22S,25R)-22,25,26-trihydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphahexacosan-19-yl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zuo, H, Park, J, Frangaj, A, Ye, J, Lu, G, Manning, J.J, Asher, W.B, Lu, Z, Hu, G, Wang, L, Mendez, J, Eng, E, Zhang, Z, Lin, X, Grasucci, R, Hendrickson, W.A, Clarke, O.B, Javitch, J.A, Conigrave, A.D, Fan, Q.R. | | Deposit date: | 2024-03-02 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Promiscuous G-protein activation by the calcium-sensing receptor.
Nature, 629, 2024
|
|
9AVL
 
 | | Structure of human calcium-sensing receptor in complex with Gi3 protein in nanodiscs | | Descriptor: | (19R,22S,25R)-22,25,26-trihydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphahexacosan-19-yl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zuo, H, Park, J, Frangaj, A, Ye, J, Lu, G, Manning, J.J, Asher, W.B, Lu, Z, Hu, G, Wang, L, Mendez, J, Eng, E, Zhang, Z, Lin, X, Grasucci, R, Hendrickson, W.A, Clarke, O.B, Javitch, J.A, Conigrave, A.D, Fan, Q.R. | | Deposit date: | 2024-03-04 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Promiscuous G-protein activation by the calcium-sensing receptor.
Nature, 629, 2024
|
|
9AXF
 
 | | Structure of human calcium-sensing receptor in complex with chimeric Gq (miniGisq) protein in detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-chlorophenyl)-N-[(1R)-1-(3-methoxyphenyl)ethyl]propan-1-amine, ... | | Authors: | Zuo, H, Park, J, Frangaj, A, Ye, J, Lu, G, Manning, J.J, Asher, W.B, Lu, Z, Hu, G, Wang, L, Mendez, J, Eng, E, Zhang, Z, Lin, X, Grasucci, R, Hendrickson, W.A, Clarke, O.B, Javitch, J.A, Conigrave, A.D, Fan, Q.R. | | Deposit date: | 2024-03-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Promiscuous G-protein activation by the calcium-sensing receptor.
Nature, 629, 2024
|
|
9AYF
 
 | | Structure of human calcium-sensing receptor in complex with Gi1 (miniGi1) protein in detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-chlorophenyl)-N-[(1R)-1-(3-methoxyphenyl)ethyl]propan-1-amine, ... | | Authors: | Zuo, H, Park, J, Frangaj, A, Ye, J, Lu, G, Manning, J.J, Asher, W.B, Lu, Z, Hu, G, Wang, L, Mendez, J, Eng, E, Zhang, Z, Lin, X, Grasucci, R, Hendrickson, W.A, Clarke, O.B, Javitch, J.A, Conigrave, A.D, Fan, Q.R. | | Deposit date: | 2024-03-07 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Promiscuous G-protein activation by the calcium-sensing receptor.
Nature, 629, 2024
|
|
7E5R
 
 | | SARS-CoV-2 S trimer with three-antibody cocktail complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FC05 heavy chain, FC05 light chain, ... | | Authors: | Sun, Y, Wang, L, Wang, N, Feng, R, Wang, X. | | Deposit date: | 2021-02-20 | | Release date: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure-based development of three- and four-antibody cocktails against SARS-CoV-2 via multiple mechanisms.
Cell Res., 31, 2021
|
|
7LJC
 
 | | Allosteric modulator LY3154207 binding to SKF-81297-bound dopamine receptor 1 in complex with miniGs protein | | Descriptor: | (1R)-6-chloro-1-phenyl-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, 2-[2,6-bis(chloranyl)phenyl]-1-[(1S,3R)-3-(hydroxymethyl)-1-methyl-5-(3-methyl-3-oxidanyl-butyl)-3,4-dihydro-1H-isoquinolin-2-yl]ethanone, CHOLESTEROL, ... | | Authors: | Zhuang, Y, Krumm, B, Zhang, H, Zhou, X.E, Wang, Y, Guo, J, Huang, X.-P, Liu, Y, Wang, L, Cheng, X, Jiang, Y, Jiang, H, Melcher, K, Zhang, C, Yi, W, Roth, B.L, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-01-28 | | Release date: | 2021-03-03 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of dopamine binding and allosteric modulation of the human D1 dopamine receptor.
Cell Res., 31, 2021
|
|
7LJD
 
 | | Allosteric modulator LY3154207 binding to dopamine-bound dopamine receptor 1 in complex with miniGs protein | | Descriptor: | 2-[2,6-bis(chloranyl)phenyl]-1-[(1S,3R)-3-(hydroxymethyl)-1-methyl-5-(3-methyl-3-oxidanyl-butyl)-3,4-dihydro-1H-isoquinolin-2-yl]ethanone, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Zhuang, Y, Krumm, B, Zhang, H, Zhou, X.E, Wang, Y, Guo, J, Huang, X.-P, Liu, Y, Wang, L, Cheng, X, Jiang, Y, Jiang, H, Melcher, K, Zhang, C, Yi, W, Roth, B.L, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-01-28 | | Release date: | 2021-03-03 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of dopamine binding and allosteric modulation of the human D1 dopamine receptor.
Cell Res., 31, 2021
|
|
3QTC
 
 | | Crystal structure of the catalytic domain of MmOmeRS, an O-methyl tyrosyl-tRNA synthetase evolved from Methanosarcina mazei PylRS, complexed with O-methyl tyrosine and AMP-PNP | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, O-methyl-L-tyrosine, ... | | Authors: | Dellas, N, Takimoto, J.K, Noel, J.P, Wang, L. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Stereochemical Basis for Engineered Pyrrolysyl-tRNA Synthetase and the Efficient in Vivo Incorporation of Structurally Divergent Non-native Amino Acids.
Acs Chem.Biol., 6, 2011
|
|
8HP5
 
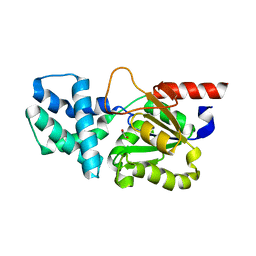 | | Crystal structure of (S)-2-haloacid dehalogenase | | Descriptor: | (S)-2-haloacid dehalogenase, 1,2-ETHANEDIOL | | Authors: | Yang, Q, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-12-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enzymatic hydrolysis on L-azetidine-2-carboxylate ring opening
Catalysis Science And Technology, 2023
|
|
8HP7
 
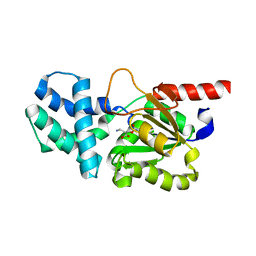 | | Crystal structure of (S)-2-haloacid dehalogenase K152A mutant trapped with (2R)-4-amino-2-hydroxybutanoic acid | | Descriptor: | (S)-2-haloacid dehalogenase, 1,2-ETHANEDIOL, GAMMA-AMINO-BUTANOIC ACID | | Authors: | Yang, Q, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-12-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Enzymatic hydrolysis on L-azetidine-2-carboxylate ring opening
Catalysis Science And Technology, 2023
|
|
8HP6
 
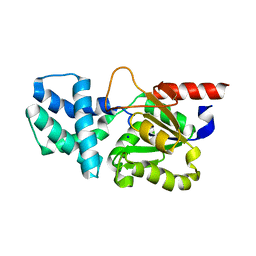 | | Crystal structure of (S)-2-haloacid dehalogenase D12A mutant | | Descriptor: | (S)-2-haloacid dehalogenase, SODIUM ION | | Authors: | Yang, Q, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-12-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enzymatic hydrolysis on L-azetidine-2-carboxylate ring opening
Catalysis Science And Technology, 2023
|
|
4JI2
 
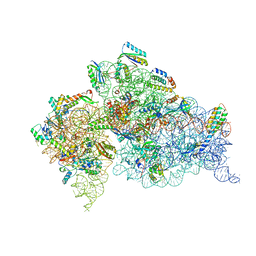 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, MAGNESIUM ION, RIBOSOMAL PROTEIN S10, ... | | Authors: | Demirci, H, Wang, L, Murphy IV, F, Murphy, E, Carr, J, Blanchard, S, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | Deposit date: | 2013-03-05 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | The central role of protein S12 in organizing the structure of the decoding site of the ribosome.
Rna, 19, 2013
|
|
4JI6
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, MAGNESIUM ION, RIBOSOMAL PROTEIN S10, ... | | Authors: | Demirci, H, Wang, L, Murphy IV, F, Murphy, E, Carr, J, Blanchard, S, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | Deposit date: | 2013-03-05 | | Release date: | 2013-11-06 | | Last modified: | 2013-12-04 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | The central role of protein S12 in organizing the structure of the decoding site of the ribosome.
Rna, 19, 2013
|
|
4JI4
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, MAGNESIUM ION, RIBOSOMAL PROTEIN S10, ... | | Authors: | Demirci, H, Wang, L, Murphy IV, F, Murphy, E, Carr, J, Blanchard, S, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | Deposit date: | 2013-03-05 | | Release date: | 2013-11-06 | | Last modified: | 2013-12-04 | | Method: | X-RAY DIFFRACTION (3.692 Å) | | Cite: | The central role of protein S12 in organizing the structure of the decoding site of the ribosome.
Rna, 19, 2013
|
|
4JI8
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, MAGNESIUM ION, RIBOSOMAL PROTEIN S10, ... | | Authors: | Demirci, H, Wang, L, Murphy IV, F, Murphy, E, Carr, J, Blanchard, S, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | Deposit date: | 2013-03-05 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.742 Å) | | Cite: | The central role of protein S12 in organizing the structure of the decoding site of the ribosome.
Rna, 19, 2013
|
|
4JI0
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, MAGNESIUM ION, ZINC ION, ... | | Authors: | Demirci, H, Wang, L, Murphy IV, F, Murphy, E, Carr, J, Blanchard, S, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | Deposit date: | 2013-03-05 | | Release date: | 2013-11-06 | | Last modified: | 2013-12-04 | | Method: | X-RAY DIFFRACTION (3.492 Å) | | Cite: | The central role of protein S12 in organizing the structure of the decoding site of the ribosome.
Rna, 19, 2013
|
|
4JI1
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, MAGNESIUM ION, RIBOSOMAL PROTEIN S10, ... | | Authors: | Demirci, H, Wang, L, Murphy IV, F, Murphy, E, Carr, J, Blanchard, S, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | Deposit date: | 2013-03-05 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.144 Å) | | Cite: | The central role of protein S12 in organizing the structure of the decoding site of the ribosome.
Rna, 19, 2013
|
|
4JI5
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, MAGNESIUM ION, RIBOSOMAL PROTEIN S10, ... | | Authors: | Demirci, H, Wang, L, Murphy IV, F, Murphy, E, Carr, J, Blanchard, S, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | Deposit date: | 2013-03-05 | | Release date: | 2013-11-06 | | Last modified: | 2013-12-04 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | The central role of protein S12 in organizing the structure of the decoding site of the ribosome.
Rna, 19, 2013
|
|
4JI7
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, MAGNESIUM ION, RIBOSOMAL PROTEIN S10, ... | | Authors: | Demirci, H, Wang, L, Murphy IV, F, Murphy, E, Carr, J, Blanchard, S, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | Deposit date: | 2013-03-05 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The central role of protein S12 in organizing the structure of the decoding site of the ribosome.
Rna, 19, 2013
|
|
4JI3
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, MAGNESIUM ION, RIBOSOMAL PROTEIN S10, ... | | Authors: | Demirci, H, Wang, L, Murphy, F.V, Murphy, E.L, Carr, J, Blanchard, S, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | Deposit date: | 2013-03-05 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | The central role of protein S12 in organizing the structure of the decoding site of the ribosome.
Rna, 19, 2013
|
|
5HGI
 
 | | Crystal structure of apo human IRE1 alpha | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BETA-MERCAPTOETHANOL, CESIUM ION, ... | | Authors: | Feldman, H.C, Tong, M, Wang, L, Meza-Acevedo, R, Gobillot, T.A, Gliedt, J.M, Hari, S.B, Mitra, A.K, Backes, B.J, Papa, F.R, Seeliger, M.A, Maly, D.J. | | Deposit date: | 2016-01-08 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.584 Å) | | Cite: | Structural and Functional Analysis of the Allosteric Inhibition of IRE1 alpha with ATP-Competitive Ligands.
Acs Chem.Biol., 11, 2016
|
|
