6WFZ
 
 | |
6WFY
 
 | |
6XC4
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CC12.3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.3 heavy chain, CC12.3 light chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
1TIM
 
 | | STRUCTURE OF TRIOSE PHOSPHATE ISOMERASE FROM CHICKEN MUSCLE | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | Banner, D.W, Bloomer, A.C, Petsko, G.A, Phillips, D.C, Wilson, I.A. | | Deposit date: | 1976-09-01 | | Release date: | 1976-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic coordinates for triose phosphate isomerase from chicken muscle.
Biochem.Biophys.Res.Commun., 72, 1976
|
|
8SDG
 
 | |
8SDF
 
 | |
8SD4
 
 | | Crystal structure of the A/Puerto Rico/8/1934 (H1N1) influenza virus hemagglutinin in complex with small molecule fusion inhibitor compound 7 | | Descriptor: | (S~1~S)-N-{3,5-dichloro-4-[(2S)-2-phenylmorpholine-4-carbonyl]phenyl}-3-[(dimethylamino)methyl]azetidine-1-sulfonimidoyl fluoride, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Kadam, R.U, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-04-06 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Ultrapotent influenza hemagglutinin fusion inhibitors developed through SuFEx-enabled high-throughput medicinal chemistry.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SD2
 
 | | Crystal structure of the A/Puerto Rico/8/1934 (H1N1) influenza virus hemagglutinin in complex with small molecule fusion inhibitor compound 4 | | Descriptor: | (S~6~S)-N-{3-chloro-4-[(2S)-2-phenylmorpholine-4-carbonyl]phenyl}-5,7-dihydro-6H-pyrrolo[3,4-b]pyridine-6-sulfonimidoyl fluoride, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kadam, R.U, Zhu, X.Y, Wilson, I.A. | | Deposit date: | 2023-04-06 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Ultrapotent influenza hemagglutinin fusion inhibitors developed through SuFEx-enabled high-throughput medicinal chemistry.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SDH
 
 | |
3CYE
 
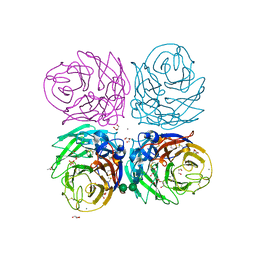 | | Cyrstal structure of the native 1918 H1N1 neuraminidase from a crystal with lattice-translocation defects | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Zhu, X, Xu, X, Wilson, I.A. | | Deposit date: | 2008-04-25 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure determination of the 1918 H1N1 neuraminidase from a crystal with lattice-translocation defects
Acta Crystallogr.,Sect.D, 64, 2008
|
|
8TJ8
 
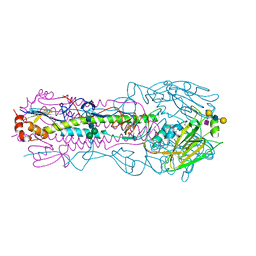 | | CRYSTAL STRUCTURE OF THE A/Moscow/10/1999(H3N2) INFLUENZA VIRUS HEMAGGLUTININ WITH HUMAN RECEPTOR ANALOG 6'-SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32, 2024
|
|
8TJ4
 
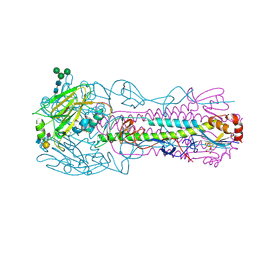 | | CRYSTAL STRUCTURE OF THE A/Bangkok/1/1979(H3N2) INFLUENZA VIRUS HEMAGGLUTININ WITH HUMAN RECEPTOR ANALOG 6'-SLNLN | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32, 2024
|
|
8TJ7
 
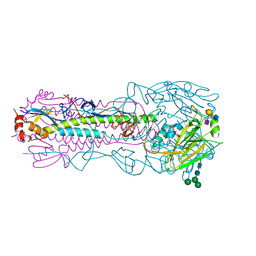 | | CRYSTAL STRUCTURE OF THE A/Shandong/9/1993(H3N2) INFLUENZA VIRUS HEMAGGLUTININ WITH HUMAN RECEPTOR ANALOG 6'-SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32, 2024
|
|
8TJB
 
 | | CRYSTAL STRUCTURE OF THE A/Texas/73/2017(H3N2) INFLUENZA VIRUS HEMAGGLUTININ WITH HUMAN RECEPTOR ANALOG 6'-SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32, 2024
|
|
8TJ6
 
 | | CRYSTAL STRUCTURE OF THE A/Beijing/353/1989(H3N2) INFLUENZA VIRUS HEMAGGLUTININ WITH HUMAN RECEPTOR ANALOG 6'-SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32, 2024
|
|
8TJ9
 
 | | CRYSTAL STRUCTURE OF THE A/Michigan/15/2014(H3N2) INFLUENZA VIRUS HEMAGGLUTININ WITH HUMAN RECEPTOR ANALOG 6'-SLNLN | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32, 2024
|
|
8TJA
 
 | | CRYSTAL STRUCTURE OF THE A/Ecuador/1374/2016(H3N2) INFLUENZA VIRUS HEMAGGLUTININ WITH HUMAN RECEPTOR ANALOG 6'-SLNLN | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32, 2024
|
|
8TGO
 
 | | Crystal structure of the BG505 triple tandem trimer gp140 HIV-1 Env in complex with PGT124 and 35O22 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 scFv, ... | | Authors: | Xian, Y, Yuan, M, Wilson, I.A. | | Deposit date: | 2023-07-12 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (5.75 Å) | | Cite: | Triple tandem trimer immunogens for HIV-1 and influenza nucleic acid-based vaccines.
Npj Vaccines, 9, 2024
|
|
8TMY
 
 | |
8TMZ
 
 | |
7LKA
 
 | | Crystal structure of SARS-CoV-2 RBD-targeting antibody COV107-23 | | Descriptor: | ACETATE ION, COV107-23 heavy chain, COV107-23 light chain | | Authors: | Yuan, M, Zhu, X, Wilson, I.A, Wu, N.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Sequence signatures of two public antibody clonotypes that bind SARS-CoV-2 receptor binding domain.
Nat Commun, 12, 2021
|
|
7LK9
 
 | | Crystal structure of SARS-CoV-2 RBD-targeting antibody COV107-23 HC + COVD21-C8 LC | | Descriptor: | COV107-23 heavy chain, COVD21-C8 light chain | | Authors: | Yuan, M, Zhu, X, Wilson, I.A, Wu, N.C. | | Deposit date: | 2021-02-01 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Sequence signatures of two public antibody clonotypes that bind SARS-CoV-2 receptor binding domain.
Nat Commun, 12, 2021
|
|
1DBA
 
 | |
1GGI
 
 | | CRYSTAL STRUCTURE OF AN HIV-1 NEUTRALIZING ANTIBODY 50.1 IN COMPLEX WITH ITS V3 LOOP PEPTIDE ANTIGEN | | Descriptor: | HIV-1 V3 LOOP PEPTIDE ANTIGEN, IGG2A 50.1 FAB (HEAVY CHAIN), IGG2A 50.1 FAB (LIGHT CHAIN) | | Authors: | Stanfield, R.L, Rini, J.M, Wilson, I.A. | | Deposit date: | 1993-04-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a human immunodeficiency virus type 1 neutralizing antibody, 50.1, in complex with its V3 loop peptide antigen.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
1GGC
 
 | |
