7BTF
 
 | | SARS-CoV-2 RNA-dependent RNA polymerase in complex with cofactors in reduced condition | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase, ... | | Authors: | Gao, Y, Yan, L, Huang, Y, Liu, F, Cao, L, Wang, T, Wang, Q, Lou, Z, Rao, Z. | | Deposit date: | 2020-04-01 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure of the RNA-dependent RNA polymerase from COVID-19 virus.
Science, 368, 2020
|
|
7DEU
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody scFv | | Authors: | Zhang, Z, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-05 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
8JAC
 
 | | Crystal structure of Mycobacterium tuberculosis LpqY in complex with trehalose analogue YB-16 | | Descriptor: | N-[[(2R,3S,4S,5R,6S)-3,4,5,6-tetrakis(oxidanyl)oxan-2-yl]methyl]ethanamide, SULFATE ION, Trehalose-binding lipoprotein LpqY, ... | | Authors: | Zhang, B, Liang, J, Rao, Z. | | Deposit date: | 2023-05-05 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular recognition of trehalose and trehalose analogues by Mycobacterium tuberculosis LpqY-SugABC.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JA7
 
 | | Cryo-EM structure of Mycobacterium tuberculosis LpqY-SugABC in complex with trehalose | | Descriptor: | Trehalose import ATP-binding protein SugC, Trehalose transport system permease protein SugA, Trehalose transport system permease protein SugB, ... | | Authors: | Zhang, B, Liang, J, Rao, Z. | | Deposit date: | 2023-05-05 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Molecular recognition of trehalose and trehalose analogues by Mycobacterium tuberculosis LpqY-SugABC.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAB
 
 | | Crystal structure of Mycobacterium tuberculosis LpqY in complex with trehalose analogue YB-06 | | Descriptor: | (2~{S},3~{S},4~{R},5~{S},6~{R})-2-(fluoranylmethyl)-6-[(2~{R},3~{R},4~{S},5~{S},6~{S})-6-(fluoranylmethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-oxane-3,4,5-triol, SULFATE ION, Trehalose-binding lipoprotein LpqY | | Authors: | Zhang, B, Liang, J, Rao, Z. | | Deposit date: | 2023-05-05 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular recognition of trehalose and trehalose analogues by Mycobacterium tuberculosis LpqY-SugABC.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAD
 
 | | Crystal structure of Mycobacterium tuberculosis LpqY in complex with trehalose analogue YB-17 | | Descriptor: | BENZOIC ACID, SULFATE ION, Trehalose-binding lipoprotein LpqY, ... | | Authors: | Zhang, B, Liang, J, Rao, Z. | | Deposit date: | 2023-05-05 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular recognition of trehalose and trehalose analogues by Mycobacterium tuberculosis LpqY-SugABC.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JA9
 
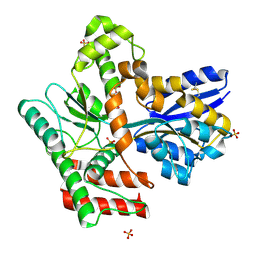 | | Crystal structure of Mycobacterium tuberculosis LpqY in complex with trehalose analogue YB-03 | | Descriptor: | SULFATE ION, Trehalose-binding lipoprotein LpqY, alpha-D-glucopyranose-(1-1)-(2~{S},3~{R},4~{S},5~{S},6~{S})-6-[(2-azanylhydrazinyl)methyl]oxane-2,3,4,5-tetrol | | Authors: | Zhang, B, Liang, J, Rao, Z. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular recognition of trehalose and trehalose analogues by Mycobacterium tuberculosis LpqY-SugABC.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAA
 
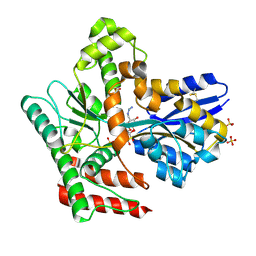 | | Crystal structure of Mycobacterium tuberculosis LpqY in complex with trehalose analogue YB-04 | | Descriptor: | (2~{S},3~{R},4~{S},5~{S},6~{S})-6-[(2-azanylhydrazinyl)methyl]oxane-2,3,4,5-tetrol, SULFATE ION, Trehalose-binding lipoprotein LpqY | | Authors: | Zhang, B, Liang, J, Rao, Z. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular recognition of trehalose and trehalose analogues by Mycobacterium tuberculosis LpqY-SugABC.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JCN
 
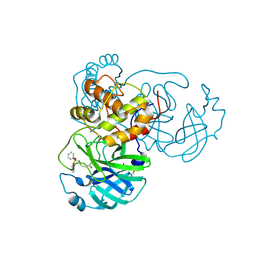 | |
8JCM
 
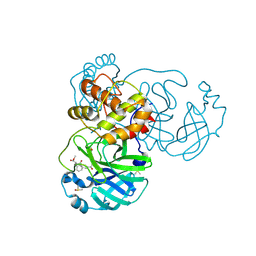 | |
8JCK
 
 | |
8JCL
 
 | |
8JCO
 
 | |
8JCJ
 
 | |
7CYQ
 
 | | Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Helicase, MAGNESIUM ION, ... | | Authors: | Yan, L, Ge, J, Zheng, L, Zhang, Y, Gao, Y, Wang, T, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM Structure of an Extended SARS-CoV-2 Replication and Transcription Complex Reveals an Intermediate State in Cap Synthesis.
Cell, 184, 2021
|
|
7CXN
 
 | | Architecture of a SARS-CoV-2 mini replication and transcription complex | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L, Zhang, Y, Ge, J, Zheng, L, Gao, Y, Wang, T, Jia, Z, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-02 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Architecture of a SARS-CoV-2 mini replication and transcription complex.
Nat Commun, 11, 2020
|
|
7CXM
 
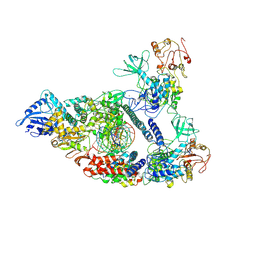 | | Architecture of a SARS-CoV-2 mini replication and transcription complex | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L, Zhang, Y, Ge, J, Zheng, L, Gao, Y, Wang, T, Jia, Z, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-02 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Architecture of a SARS-CoV-2 mini replication and transcription complex.
Nat Commun, 11, 2020
|
|
7DET
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody scFv | | Authors: | Wang, Y, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-05 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
8JA8
 
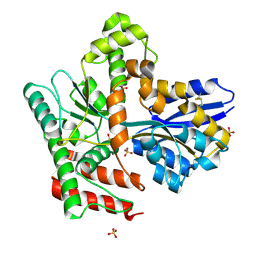 | |
7YVN
 
 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH281 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH281 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVP
 
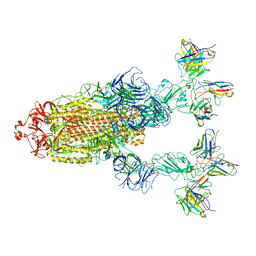 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH272/281 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH272 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVG
 
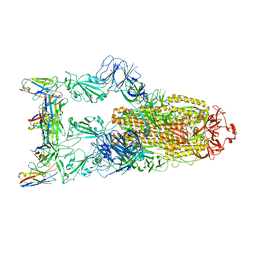 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH132 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH132 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVK
 
 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH272 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH272 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVO
 
 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH027/132 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH132 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVH
 
 | | Omicron BA.4/5 SARS-CoV-2 S RBD in complex with TH132 Fab | | Descriptor: | Spike glycoprotein, TH132 Fab heavy chain, TH132 Fab light chain | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
