7EXV
 
 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with beta-L-arabinofuranoylamide | | Descriptor: | 2-bromanyl-N-[(2S,3R,4R,5S)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]ethanamide, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Sawano, K, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Substrate complex structure, active site labeling and catalytic role of the zinc ion in cysteine glycosidase.
Glycobiology, 32, 2022
|
|
5YIZ
 
 | |
5YJ0
 
 | |
5YJ1
 
 | | Mouse Cereblon thalidomide binding domain complexed with R-form thalidomide | | Descriptor: | 2-[(3~{R})-2,6-bis(oxidanylidene)piperidin-3-yl]isoindole-1,3-dione, Protein cereblon, SULFATE ION, ... | | Authors: | Mori, T, Hakoshima, T. | | Deposit date: | 2017-10-06 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of thalidomide enantiomer binding to cereblon
Sci Rep, 8, 2018
|
|
7DIF
 
 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with beta-L-arabinofuranose-configured cyclophellitol at 1.75-angstrom resolution | | Descriptor: | (1S,2S,3R,4R)-3-(hydroxymethyl)cyclopentane-1,2,4-triol, Non-reducing end beta-L-arabinofuranosidase, POTASSIUM ION, ... | | Authors: | Amaki, S, McGregor, N.G.S, Arakawa, T, Yamada, C, Borlandelli, V, Overkleeft, H.S, Davies, G.J, Fushinobu, S. | | Deposit date: | 2020-11-19 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cysteine Nucleophiles in Glycosidase Catalysis: Application of a Covalent beta-l-Arabinofuranosidase Inhibitor.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
2RQR
 
 | | The solution structure of human DOCK2 SH3 domain - ELMO1 peptide chimera complex | | Descriptor: | Engulfment and cell motility protein 1,Dedicator of cytokinesis protein 2 | | Authors: | Yokoyama, S, Tochio, N, Koshiba, S, Kigawa, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-10-21 | | Release date: | 2010-10-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for mutual relief of the Rac guanine nucleotide exchange factor DOCK2 and its partner ELMO1 from their autoinhibited forms.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6KAZ
 
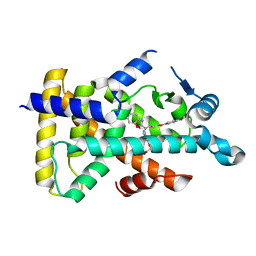 | | X-ray structure of human PPARalpha ligand binding domain-pemafibrate co-crystals obtained by soaking | | Descriptor: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KB5
 
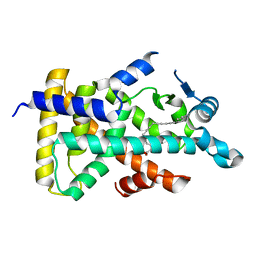 | | X-ray structure of human PPARalpha ligand binding domain-5,8,11,14-eicosatetraynoic Acid (ETYA) co-crystals obtained by delipidation and cross-seeding | | Descriptor: | GLYCEROL, Peroxisome proliferator-activated receptor alpha, icosa-5,8,11,14-tetraynoic acid | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KBA
 
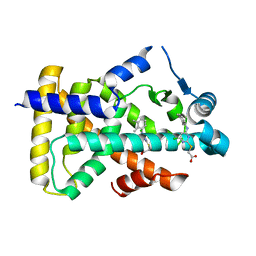 | | X-ray structure of human PPARalpha ligand binding domain-Wy14643 co-crystals obtained by co-crystallization | | Descriptor: | 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KAX
 
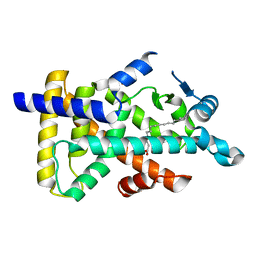 | | X-ray structure of human PPARalpha ligand binding domain-intrinsic fatty acid (E. coli origin) co-crystals obtained by cross-seeding | | Descriptor: | GLYCEROL, PALMITIC ACID, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KB4
 
 | | X-ray structure of human PPARalpha ligand binding domain-pemafibrate co-crystals obtained by delipidation and cross-seeding | | Descriptor: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KB6
 
 | | X-ray structure of human PPARalpha ligand binding domain-tetradecylthioacetic acid (TTA) co-crystals obtained by delipidation and cross-seeding | | Descriptor: | 2-tetradecylsulfanylethanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.431 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KYP
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-clofibric acid co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-(4-chloranylphenoxy)-2-methyl-propanoic acid, 2-chloro-5-nitro-N-phenylbenzamide, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-09-19 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6L37
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-ciprofibrate co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-chloro-5-nitro-N-phenylbenzamide, 2-{4-[(1S)-2,2-dichlorocyclopropyl]phenoxy}-2-methylpropanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-10-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6L36
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-fenofibric acid co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-[4-(4-chlorobenzene-1-carbonyl)phenoxy]-2-methylpropanoic acid, 2-chloro-5-nitro-N-phenylbenzamide, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-10-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6L38
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-gemfibrozil co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-chloro-5-nitro-N-phenylbenzamide, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-10-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.761 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
3B13
 
 | | Crystal structure of the DHR-2 domain of DOCK2 in complex with Rac1 (T17N mutant) | | Descriptor: | Dedicator of cytokinesis protein 2, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Hanawa-Suetsugu, K, Kukimoto-Niino, M, Mishima-Tsumagari, C, Terada, T, Shirouzu, M, Fukui, Y, Yokoyama, S. | | Deposit date: | 2011-06-24 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.006 Å) | | Cite: | Structural basis for mutual relief of the Rac guanine nucleotide exchange factor DOCK2 and its partner ELMO1 from their autoinhibited forms.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7EBG
 
 | | Crystal structure of human pyruvate dehydrogenase kinase 4 in complex with compound 7 | | Descriptor: | 3,3-dimethyl-7-(methylamino)-1H-indol-2-one, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Orita, T, Doi, S, Iwanaga, T, Adachi, T. | | Deposit date: | 2021-03-09 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment-based lead discovery to identify novel inhibitors that target the ATP binding site of pyruvate dehydrogenase kinases.
Bioorg.Med.Chem., 44, 2021
|
|
7EAT
 
 | | Crystal structure of human pyruvate dehydrogenase kinase 4 in complex with compound 1 | | Descriptor: | 1,3-dihydro-2H-indol-2-one, SULFATE ION, [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 4, ... | | Authors: | Orita, T, Doi, S, Iwanaga, T, Adachi, T. | | Deposit date: | 2021-03-08 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fragment-based lead discovery to identify novel inhibitors that target the ATP binding site of pyruvate dehydrogenase kinases.
Bioorg.Med.Chem., 44, 2021
|
|
7EBB
 
 | | Crystal structure of human pyruvate dehydrogenase kinase 4 in complex with compound 2 | | Descriptor: | 1H-pyrrolo[2,3-b]pyridine-3-carbonitrile, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Orita, T, Doi, S, Iwanaga, T, Adachi, T. | | Deposit date: | 2021-03-09 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-based lead discovery to identify novel inhibitors that target the ATP binding site of pyruvate dehydrogenase kinases.
Bioorg.Med.Chem., 44, 2021
|
|
7EA0
 
 | | Crystal structure of human pyruvate dehydrogenase kinase 2 in complex with compound 1 | | Descriptor: | 1,3-dihydro-2H-indol-2-one, ACETATE ION, CHLORIDE ION, ... | | Authors: | Orita, T, Doi, S, Iwanaga, T, Adachi, T. | | Deposit date: | 2021-03-05 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Fragment-based lead discovery to identify novel inhibitors that target the ATP binding site of pyruvate dehydrogenase kinases.
Bioorg.Med.Chem., 44, 2021
|
|
7EAS
 
 | | Crystal structure of human pyruvate dehydrogenase kinase 2 in complex with compound 2 | | Descriptor: | 1H-pyrrolo[2,3-b]pyridine-3-carbonitrile, ACETATE ION, CHLORIDE ION, ... | | Authors: | Orita, T, Doi, S, Iwanaga, T, Adachi, T. | | Deposit date: | 2021-03-08 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Fragment-based lead discovery to identify novel inhibitors that target the ATP binding site of pyruvate dehydrogenase kinases.
Bioorg.Med.Chem., 44, 2021
|
|
7EBH
 
 | | Crystal structure of human pyruvate dehydrogenase kinase 2 in complex with compound 13 | | Descriptor: | 5-bromanyl-2-methyl-6-propyl-7H-pyrrolo[2,3-d]pyrimidine, ACETATE ION, CHLORIDE ION, ... | | Authors: | Orita, T, Doi, S, Iwanaga, T, Adachi, T. | | Deposit date: | 2021-03-09 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Fragment-based lead discovery to identify novel inhibitors that target the ATP binding site of pyruvate dehydrogenase kinases.
Bioorg.Med.Chem., 44, 2021
|
|
4YOT
 
 | |
4YOU
 
 | |
