6O2T
 
 | | Acetylated Microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eshun-Wilson, L, Zhang, R, Portran, D, Nachury, M.V, Toso, D, Lohr, T, Vendruscolo, M, Bonomi, M, Fraser, J.S, Nogales, E. | | Deposit date: | 2019-02-24 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Effects of alpha-tubulin acetylation on microtubule structure and stability.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6DLM
 
 | | DHD127 | | Descriptor: | DHD127_A, DHD127_B | | Authors: | Bick, M.J, Chen, Z, Baker, D. | | Deposit date: | 2018-06-01 | | Release date: | 2018-12-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.753 Å) | | Cite: | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
6O2S
 
 | | Deacetylated Microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eshun-Wilson, L, Zhang, R, Portran, D, Nachury, M.V, Toso, D, Lohr, T, Vendruscolo, M, Bonomi, M, Fraser, J.S, Nogales, E. | | Deposit date: | 2019-02-24 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Effects of alpha-tubulin acetylation on microtubule structure and stability.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6O2R
 
 | | Deacetylated Microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eshun-Wilson, L, Zhang, R, Portran, D, Nachury, M.V, Toso, D, Lohr, T, Vendruscolo, M, Bonomi, M, Fraser, J.S, Nogales, E. | | Deposit date: | 2019-02-24 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Effects of alpha-tubulin acetylation on microtubule structure and stability.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3MYQ
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 2-chloro-5-[(1H-imidazo[4,5-c]quinolin-2-ylsulfanyl)acetyl]benzenesulfonamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloro-5-[(1H-imidazo[4,5-c]quinolin-2-ylsulfanyl)acetyl]benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Grazulis, S, Manakova, E, Golovenko, D. | | Deposit date: | 2010-05-11 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Indapamide-like benzenesulfonamides as inhibitors of carbonic anhydrases I, II, VII, and XIII.
Bioorg.Med.Chem., 18, 2010
|
|
4CGX
 
 | | Crystal structure of the N-terminal domain of the PA subunit of Thogoto virus polymerase (form 1) | | Descriptor: | POLYMERASE ACIDIC PROTEIN | | Authors: | Guilligay, D, Kadlec, J, Crepin, T, Lunardi, T, Bouvier, D, Kochs, G, Ruigrok, R.W.H, Cusack, S. | | Deposit date: | 2013-11-27 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Comparative Structural and Functional Analysis of Orthomyxovirus Polymerase CAP-Snatching Domains.
Plos One, 9, 2014
|
|
3N52
 
 | | crystal Structure analysis of MIP2 | | Descriptor: | C-X-C motif chemokine 2 | | Authors: | Rajasekaran, D. | | Deposit date: | 2010-05-24 | | Release date: | 2011-06-08 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Model of GAG/MIP-2/CXCR2 Interfaces and Its Functional Effects.
Biochemistry, 51, 2012
|
|
4CIK
 
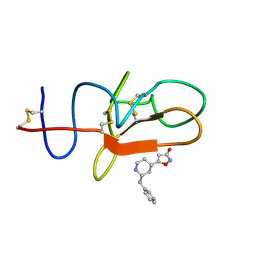 | | plasminogen kringle 1 in complex with inhibitor | | Descriptor: | 5-[(2R,4S)-2-(phenylmethyl)piperidin-4-yl]-1,2-oxazol-3-one, PLASMINOGEN | | Authors: | Xue, Y, Johansson, C, Cheng, L, Pettersen, D, Gustafsson, D. | | Deposit date: | 2013-12-10 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Discovery of the Fibrinolysis Inhibitor Azd6564, Acting Via Interference of a Protein-Protein Interaction.
Acs Med.Chem.Lett., 5, 2014
|
|
8C7G
 
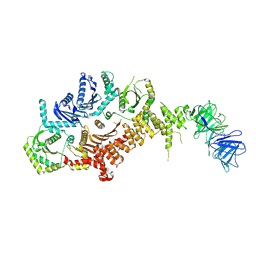 | | Drosophila melanogaster Rab7 GEF complex Mon1-Ccz1-Bulli | | Descriptor: | Caffeine, calcium, zinc sensitivity 1, ... | | Authors: | Schaefer, J, Herrmann, E, Kuemmel, D, Moeller, A. | | Deposit date: | 2023-01-15 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the metazoan Rab7 GEF complex Mon1-Ccz1-Bulli.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8CPO
 
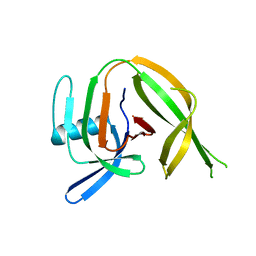 | | Crystal structure of the PolB16_OarG intein variant S1A, N183A, C111A, C165A | | Descriptor: | PolB16 Intein Cys-less | | Authors: | Kattelmann, S, Pasch, T, Mootz, H.D, Kuemmel, D. | | Deposit date: | 2023-03-03 | | Release date: | 2023-05-17 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical analysis of a novel atypically split intein reveals a conserved histidine specific to cysteine-less inteins.
Chem Sci, 14, 2023
|
|
8WQO
 
 | | Crystal structure of BRD4-BD1 bound with DI106 | | Descriptor: | (3~{R})-6-[[4-(3,5-dimethyl-1~{H}-pyrazol-4-yl)pyrimidin-2-yl]amino]-1,3-dimethyl-4-propan-2-yl-3~{H}-quinoxalin-2-one, Isoform C of Bromodomain-containing protein 4 | | Authors: | Cao, D, Xiong, B. | | Deposit date: | 2023-10-12 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Rational design and evaluation of BET-Aurora A dual-inhibitors for treatment of cancers
To Be Published
|
|
4CIS
 
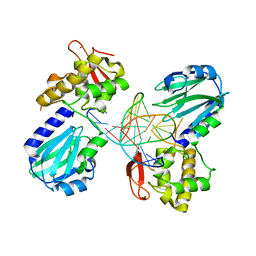 | | Structure of MutM in complex with carbocyclic 8-oxo-G containing DNA | | Descriptor: | (R,R)-2,3-BUTANEDIOL, DNA, FORMAMIDOPYRIMIDIN DNA GLYCOSYLASE, ... | | Authors: | Schneider, S, Sadeghian, K, Flaig, D, Blank, I.D, Strasser, R, Stathis, D, Winnacker, M, Carell, T, Ochsenfeld, C. | | Deposit date: | 2013-12-15 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Ribose-protonated DNA base excision repair: a combined theoretical and experimental study.
Angew. Chem. Int. Ed. Engl., 53, 2014
|
|
3NHC
 
 | | GYMLGS segment 127-132 from human prion with M129 | | Descriptor: | Major prion protein | | Authors: | Apostol, M.I, Eisenberg, D. | | Deposit date: | 2010-06-14 | | Release date: | 2010-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystallographic studies of prion protein (PrP) segments suggest how structural changes encoded by polymorphism at residue 129 modulate susceptibility to human prion disease.
J.Biol.Chem., 285, 2010
|
|
8BXT
 
 | | Structure of StayGold | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, StayGold | | Authors: | Ivorra-Molla, E, Akhuli, D, Crow, A. | | Deposit date: | 2022-12-09 | | Release date: | 2023-07-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A monomeric StayGold fluorescent protein.
Nat.Biotechnol., 2023
|
|
8CCI
 
 | | Crystal structure of Mycobacterium smegmatis thioredoxin reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fuesser, F.T, Koch, O, Kuemmel, D. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Novel starting points for fragment-based drug design against mycobacterial thioredoxin reductase identified using crystallographic fragment screening.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8XFB
 
 | | Cryo-EM structure of partial dimeric WDR11-FAM91A1 complex | | Descriptor: | Protein FAM91A1, WD repeat-containing protein 11 | | Authors: | Jia, G.W, Deng, Q.H, Su, Z.M, Jia, D. | | Deposit date: | 2023-12-13 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The WDR11 complex is a receptor for acidic-cluster-containing cargo proteins.
Cell, 187, 2024
|
|
8XI4
 
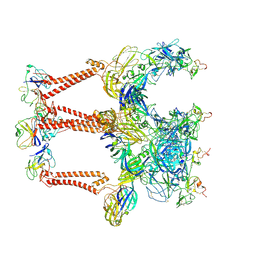 | | Structure of Eastern Equine Encephalitis VLP in complex with the receptor VLDLR LA1-2 | | Descriptor: | CALCIUM ION, Capsid protein, Spike glycoprotein E1, ... | | Authors: | Cao, D, Ma, B, Cao, Z, Xu, X, Zhang, X, Xiang, Y. | | Deposit date: | 2023-12-19 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The receptor VLDLR binds Eastern Equine Encephalitis virus through multiple distinct modes.
Nat Commun, 15, 2024
|
|
8XI5
 
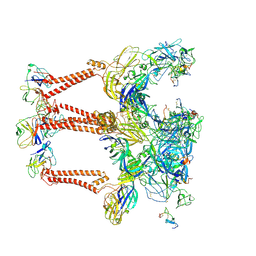 | | Structure of Eastern Equine Encephalitis VLP in complex with the receptor VLDLR LA3-5 | | Descriptor: | CALCIUM ION, Capsid protein, Spike glycoprotein E1, ... | | Authors: | Cao, D, Ma, B, Cao, Z, Xu, X, Zhang, X, Xiang, Y. | | Deposit date: | 2023-12-19 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The receptor VLDLR binds Eastern Equine Encephalitis virus through multiple distinct modes.
Nat Commun, 15, 2024
|
|
9B3P
 
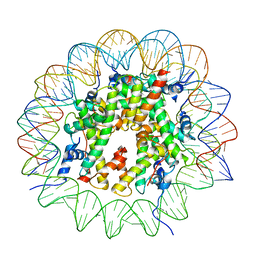 | |
8YGW
 
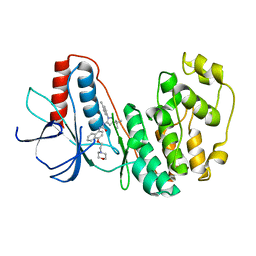 | | The Crystal Structure of MAPK11 from Biortus | | Descriptor: | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA, Mitogen-activated protein kinase 11 | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Guo, S. | | Deposit date: | 2024-02-27 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | The Crystal Structure of MAPK11 from Biortus.
To Be Published
|
|
8YK5
 
 | |
8C6C
 
 | | Light SFX structure of D.m(6-4)photolyase at 300ps time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C1U
 
 | | SFX structure of D.m(6-4)photolyase | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C6H
 
 | | Light SFX structure of D.m(6-4)photolyase at 2ps time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C69
 
 | | Light SFX structure of D.m(6-4)photolyase at 100 microsecond time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
