5CSB
 
 | | The crystal structure of beta2-microglobulin D76N mutant at room temperature | | Descriptor: | Beta-2-microglobulin | | Authors: | de Rosa, M, Mota, C.S, de Sanctis, D, Bolognesi, M, Ricagno, S. | | Deposit date: | 2015-07-23 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.719 Å) | | Cite: | Conformational dynamics in crystals reveal the molecular bases for D76N beta-2 microglobulin aggregation propensity.
Nat Commun, 9, 2018
|
|
4RMS
 
 | |
4RMT
 
 | | Crystal structure of the D98N Beta-2 Microglobulin mutant | | Descriptor: | ACETATE ION, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | de Rosa, M, Bolognesi, M, Ricagno, S. | | Deposit date: | 2014-10-22 | | Release date: | 2015-11-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.242 Å) | | Cite: | Decoding the Structural Bases of D76N 2-Microglobulin High Amyloidogenicity through Crystallography and Asn-Scan Mutagenesis.
Plos One, 10, 2015
|
|
5CS7
 
 | | The crystal structure of wt beta2-microglobulin at room temperature | | Descriptor: | Beta-2-microglobulin | | Authors: | de Rosa, M, Mota, C.S, de Sanctis, D, Bolognesi, M, Ricagno, S. | | Deposit date: | 2015-07-23 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational dynamics in crystals reveal the molecular bases for D76N beta-2 microglobulin aggregation propensity.
Nat Commun, 9, 2018
|
|
4RMW
 
 | |
8AYF
 
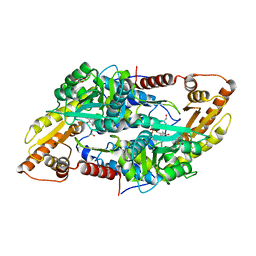 | | Crystal structure of human Sphingosine-1-phosphate lyase 1 | | Descriptor: | ACETATE ION, GLYCEROL, Sphingosine-1-phosphate lyase 1 | | Authors: | Giardina, G, Catalano, F, Pampalone, G, Cellini, B. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Dual species sphingosine-1-phosphate lyase inhibitors to combine antifungal and anti-inflammatory activities in cystic fibrosis: a feasibility study.
Sci Rep, 13, 2023
|
|
5FAF
 
 | | N184K pathological variant of gelsolin domain 2 (orthorhombic form) | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Boni, F, Milani, M, Ricagno, s, Bolognesi, M, de Rosa, M. | | Deposit date: | 2015-12-11 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Molecular basis of a novel renal amyloidosis due to N184K gelsolin variant.
Sci Rep, 6, 2016
|
|
4RMU
 
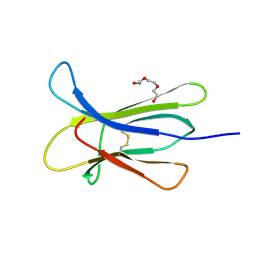 | |
4RMV
 
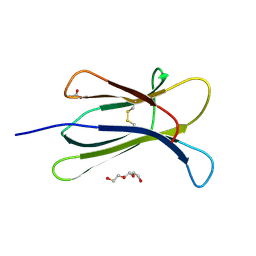 | |
4RMR
 
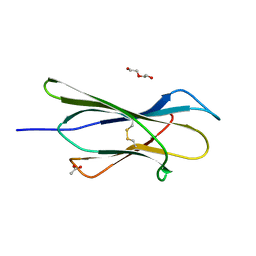 | |
4RMQ
 
 | |
4L3C
 
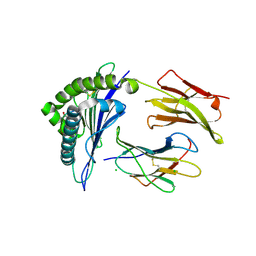 | | Structure of HLA-A2 in complex with D76N b2m mutant and NY-ESO1 double mutant | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, GLYCEROL, ... | | Authors: | Halabelian, L, Giorgetti, S, Bellotti, V, Bolognesi, M, Ricagno, S. | | Deposit date: | 2013-06-05 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Class I Major Histocompatibility Complex, the Trojan Horse for Secretion of Amyloidogenic beta 2-Microglobulin.
J.Biol.Chem., 289, 2014
|
|
5FAE
 
 | | N184K pathological variant of gelsolin domain 2 (trigonal form) | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Boni, F, Milani, M, Ricagno, S, Bolognesi, M, de Rosa, M. | | Deposit date: | 2015-12-11 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of a novel renal amyloidosis due to N184K gelsolin variant.
Sci Rep, 6, 2016
|
|
4L29
 
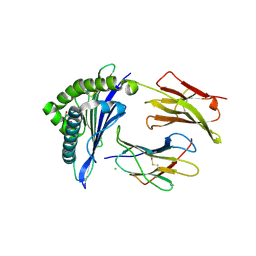 | | Structure of wtMHC class I with NY-ESO1 double mutant | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, GLYCEROL, ... | | Authors: | Halabelian, L, Giorgetti, S, Bellotti, V, Bolognesi, M, Ricagno, S. | | Deposit date: | 2013-06-04 | | Release date: | 2013-12-25 | | Last modified: | 2014-02-26 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Class I Major Histocompatibility Complex, the Trojan Horse for Secretion of Amyloidogenic beta 2-Microglobulin.
J.Biol.Chem., 289, 2014
|
|
7ZS6
 
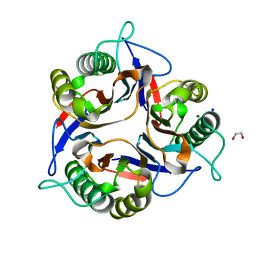 | | Crystal structure of Apis mellifera RidA | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Reactive intermediate deaminase A, ... | | Authors: | Visentin, C, Rizzi, G, Ricagno, S. | | Deposit date: | 2022-05-06 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Apis mellifera RidA, a novel member of the canonical YigF/YER057c/UK114 imine deiminase superfamily of enzymes pre-empting metabolic damage.
Biochem.Biophys.Res.Commun., 616, 2022
|
|
1GAR
 
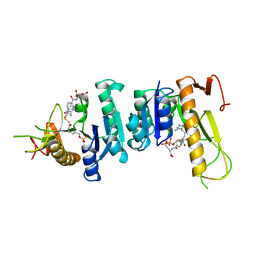 | | TOWARDS STRUCTURE-BASED DRUG DESIGN: CRYSTAL STRUCTURE OF A MULTISUBSTRATE ADDUCT COMPLEX OF GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE AT 1.96 ANGSTROMS RESOLUTION | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE, N-[4-[[3-(2,4-DIAMINO-1,6-DIHYDRO-6-OXO-4-PYRIMIDINYL)-PROPYL]-[2-((2-OXO-2-((4-PHOSPHORIBOXY)-BUTYL)-AMINO)-ETHYL)-THIO-ACETYL]-AMINO]BENZOYL]-1-GLUTAMIC ACID | | Authors: | Wilson, I.A, Klein, C, Chen, P, Arevalo, J.H. | | Deposit date: | 1994-12-08 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Towards structure-based drug design: crystal structure of a multisubstrate adduct complex of glycinamide ribonucleotide transformylase at 1.96 A resolution.
J.Mol.Biol., 249, 1995
|
|
3L1A
 
 | |
3L2Z
 
 | | Crystal structure of hydrated Biotin Protein Ligase from M. tuberculosis | | Descriptor: | BirA bifunctional protein | | Authors: | Gupta, V, Gupta, R.K, Khare, G, Salunke, D.M, Tyagi, A.K. | | Deposit date: | 2009-12-16 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural ordering of disordered ligand-binding loops of biotin protein ligase into active conformations as a consequence of dehydration.
Plos One, 5, 2010
|
|
6UEL
 
 | | CPS1 bound to allosteric inhibitor H3B-193 | | Descriptor: | Carbamoyl-phosphate synthase [ammonia], mitochondrial, N~1~-[(4-fluorophenyl)methyl]-N~1~-methyl-N~4~-(4-methyl-1,3-thiazol-2-yl)piperidine-1,4-dicarboxamide, ... | | Authors: | Larsen, N.A, Nguyen, T.V. | | Deposit date: | 2019-09-21 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small Molecule Inhibition of CPS1 Activity through an Allosteric Pocket.
Cell Chem Biol, 27, 2020
|
|
2BDN
 
 | | Crystal structure of human MCP-1 bound to a blocking antibody, 11K2 | | Descriptor: | Antibody heavy chain 11K2, Antibody light chain 11K2, Small inducible cytokine A2 | | Authors: | Boriack-Sjodin, P.A, Rushe, M, Reid, C, Jarpe, M, van Vlijmen, H, Bailly, V. | | Deposit date: | 2005-10-20 | | Release date: | 2006-06-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure activity relationships of monocyte chemoattractant proteins in complex with a blocking antibody.
Protein Eng.Des.Sel., 19, 2006
|
|
3CW1
 
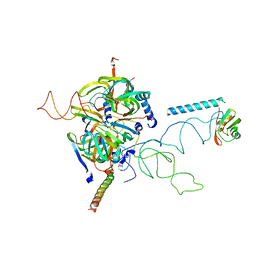 | | Crystal Structure of Human Spliceosomal U1 snRNP | | Descriptor: | Small nuclear ribonucleoprotein E, Small nuclear ribonucleoprotein F, Small nuclear ribonucleoprotein G, ... | | Authors: | Pomeranz Krummel, D.A, Oubridge, C, Leung, A.K, Li, J, Nagai, K. | | Deposit date: | 2008-04-21 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (5.493 Å) | | Cite: | Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution.
Nature, 458, 2009
|
|
4PTL
 
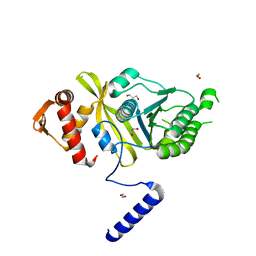 | | Mycobacterium tuberculosis RecA glycerol bound low temperature structure IIC-GM | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein RecA, ... | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-03-11 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
4OQF
 
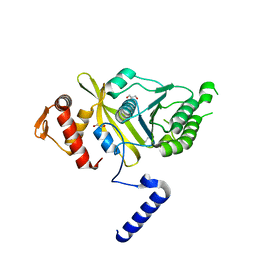 | | Mycobacterium tuberculosis RecA glycerol bound low temperature structure IIB-SR | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein RecA | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-02-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
4PPG
 
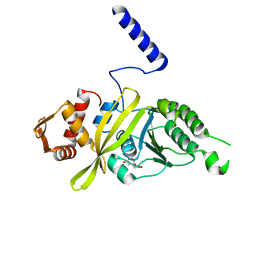 | | Mycobacterium tuberculosis RecA citrate bound low temperature structure IIA-BR | | Descriptor: | CITRATE ANION, Protein RecA, 1st part, ... | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
4PPQ
 
 | | Mycobacterium tuberculosis RecA citrate bound low temperature structure IIA-CR | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Protein RecA, ... | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
