6KEL
 
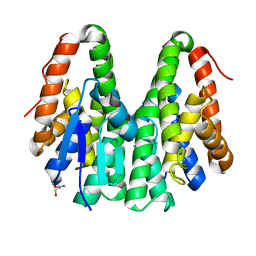 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in apo-form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutathione S-transferase E14 | | Authors: | Koiwai, K, Inaba, K, Morohashi, K, Yumoto, F, Senda, T, Niwa, R. | | Deposit date: | 2019-07-04 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An integrated approach to unravel a crucial structural property required for the function of the insect steroidogenic Halloween protein Noppera-bo.
J.Biol.Chem., 295, 2020
|
|
6KEQ
 
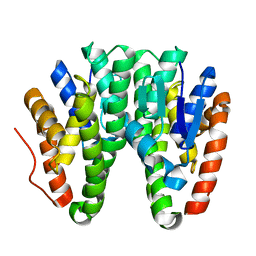 | | Crystal structure of D113A mutant of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in apo-form | | Descriptor: | Glutathione S-transferase E14 | | Authors: | Koiwai, K, Inaba, K, Morohashi, K, Yumoto, F, Niwa, R, Senda, T. | | Deposit date: | 2019-07-04 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | An integrated approach to unravel a crucial structural property required for the function of the insect steroidogenic Halloween protein Noppera-bo.
J.Biol.Chem., 295, 2020
|
|
6KEN
 
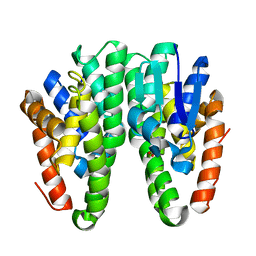 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in glutathione-bound form | | Descriptor: | GLUTATHIONE, Glutathione S-transferase E14 | | Authors: | Koiwai, K, Inaba, K, Morohashi, K, Yumoto, F, Niwa, R, Senda, T. | | Deposit date: | 2019-07-04 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An integrated approach to unravel a crucial structural property required for the function of the insect steroidogenic Halloween protein Noppera-bo.
J.Biol.Chem., 295, 2020
|
|
6LYC
 
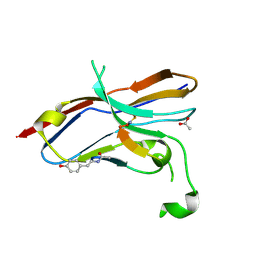 | | Crystal structure of the NOD SIRPa complex with D4-2 | | Descriptor: | ACETIC ACID, D4-2, SIRPa of the NOD mouse strain | | Authors: | Murata, Y, Matsuda, M, Nakagawa, A, Matozaki, T. | | Deposit date: | 2020-02-14 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Macrocyclic Peptide-Mediated Blockade of the CD47-SIRP alpha Interaction as a Potential Cancer Immunotherapy.
Cell Chem Biol, 27, 2020
|
|
6FWF
 
 | | Low resolution structure of Neisseria meningitidis qNOR | | Descriptor: | CALCIUM ION, FE (III) ION, Nitric-oxide reductase, ... | | Authors: | Young, D, Antonyuk, S, Tosha, T, Hisano, T, Hasnain, S, Shiro, Y. | | Deposit date: | 2018-03-06 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Characterization of the quinol-dependent nitric oxide reductase from the pathogen Neisseria meningitidis, an electrogenic enzyme.
Sci Rep, 8, 2018
|
|
7DC8
 
 | | Crystal structure of Switch Ab Fab and hIL6R in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Interleukin-6 receptor subunit alpha, SULFATE ION, ... | | Authors: | Kadono, S, Fukami, T.A, Kawauchi, H, Torizawa, T, Mimoto, F. | | Deposit date: | 2020-10-23 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.757 Å) | | Cite: | Exploitation of Elevated Extracellular ATP to Specifically Direct Antibody to Tumor Microenvironment.
Cell Rep, 33, 2020
|
|
6VL5
 
 | |
6VL6
 
 | |
6VKN
 
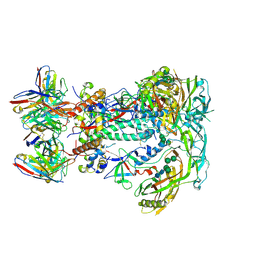 | | BG505 SOSIP.v5.2.N241.N289 in complex with rhesus macaque Fab RM19R | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cottrell, C.A, Ward, A.B. | | Deposit date: | 2020-01-21 | | Release date: | 2020-08-12 | | Last modified: | 2020-09-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Targeting HIV Env immunogens to B cell follicles in nonhuman primates through immune complex or protein nanoparticle formulations.
NPJ Vaccines, 5, 2020
|
|
5WQJ
 
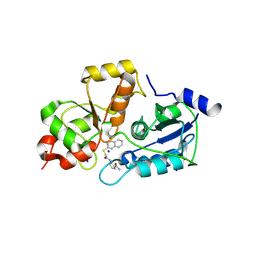 | |
5WQK
 
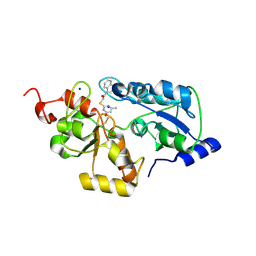 | |
6YTU
 
 | | Atomic-resolution structure of the coiled-coil dimerisation domain of human Arc | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Activity-regulated cytoskeleton-associated protein, CHLORIDE ION | | Authors: | Hallin, E.I, Touma, C, Bramham, C.R, Kursula, P. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-03 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Arc self-association and formation of virus-like capsids are mediated by an N-terminal helical coil motif.
Febs J., 288, 2021
|
|
1DZE
 
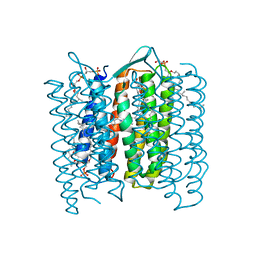 | | Structure of the M Intermediate of Bacteriorhodopsin trapped at 100K | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, 3-PHOSPHORYL-[1,2-DI-PHYTANYL]GLYCEROL, ... | | Authors: | Takeda, K, Matsui, Y, Sato, H, Hino, T, Kanamori, E, Okumura, H, Yamane, T, Iizuka, T, Kamiya, N, Adachi, S, Kouyama, T. | | Deposit date: | 2000-02-25 | | Release date: | 2000-08-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the M Intermediate of Bacteriorhodopsin: Allosteric Structural Changes Mediated by Sliding Movement of a Transmembrane Helix
J.Mol.Biol., 341, 2004
|
|
2D9Q
 
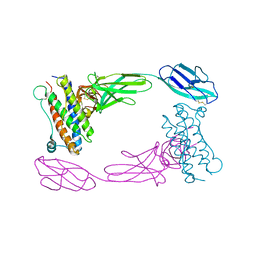 | | Crystal Structure of the Human GCSF-Receptor Signaling Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CSF3, Granulocyte colony-stimulating factor receptor | | Authors: | Tamada, T, Kuroki, R. | | Deposit date: | 2005-12-12 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Homodimeric cross-over structure of the human granulocyte colony-stimulating factor (GCSF) receptor signaling complex
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1RDV
 
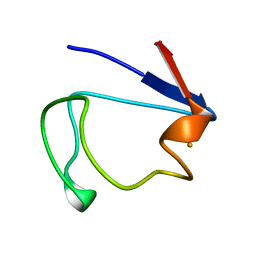 | |
3UIX
 
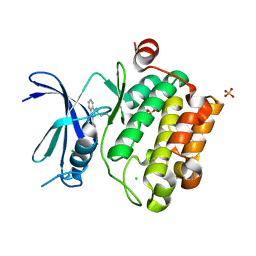 | |
2YQS
 
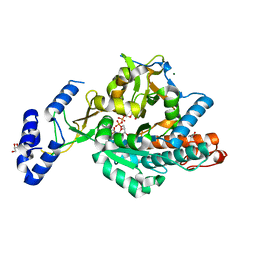 | | Crystal structure of uridine-diphospho-N-acetylglucosamine pyrophosphorylase from Candida albicans, in the product-binding form | | Descriptor: | GLYCEROL, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Miki, K, Maruyama, D, Nishitani, Y, Nonaka, T, Kita, A. | | Deposit date: | 2007-03-30 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Uridine-diphospho-N-acetylglucosamine Pyrophosphorylase from Candida albicans and Catalytic Reaction Mechanism
J.Biol.Chem., 282, 2007
|
|
2YQJ
 
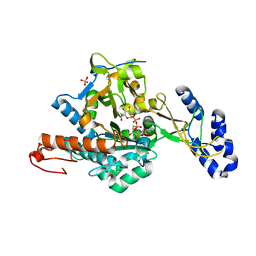 | | Crystal Structure of uridine-diphospho-N-acetylglucosamine pyrophosphorylase from Candida albicans, in the reaction-completed form | | Descriptor: | GLYCEROL, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Miki, K, Maruyama, D, Nishitani, Y, Nonaka, T, Kita, A. | | Deposit date: | 2007-03-30 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure of Uridine-diphospho-N-acetylglucosamine Pyrophosphorylase from Candida albicans and Catalytic Reaction Mechanism
J.Biol.Chem., 282, 2007
|
|
2YQC
 
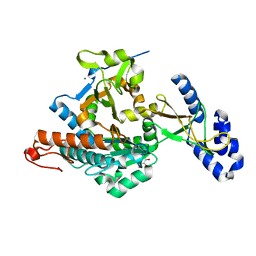 | | Crystal Structure of uridine-diphospho-N-acetylglucosamine pyrophosphorylase from Candida albicans, in the apo-like form | | Descriptor: | GLYCEROL, MAGNESIUM ION, UDP-N-acetylglucosamine pyrophosphorylase | | Authors: | Miki, K, Maruyama, D, Nishitani, Y, Nonaka, T, Kita, A. | | Deposit date: | 2007-03-30 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Uridine-diphospho-N-acetylglucosamine Pyrophosphorylase from Candida albicans and Catalytic Reaction Mechanism
J.Biol.Chem., 282, 2007
|
|
7CJ3
 
 | | Crystal structure of the transmembrane domain of Salpingoeca rosetta rhodopsin phosphodiesterase | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Phosphodiesterase, RETINAL | | Authors: | Ikuta, T, Shihoya, W, Yamashita, K, Nureki, O. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the mechanism of rhodopsin phosphodiesterase.
Nat Commun, 11, 2020
|
|
4YD9
 
 | | Crystal structure of squid hemocyanin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CU2-O2 CLUSTER, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Matsuno, A, Gai, Z, Kato, K, Tanaka, Y, Yao, M. | | Deposit date: | 2015-02-21 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the 3.8-MDa Respiratory Supermolecule Hemocyanin at 3.0 angstrom Resolution
Structure, 23, 2015
|
|
7D7Q
 
 | | Crystal structure of the transmembrane domain and linker region of Salpingoeca rosetta rhodopsin phosphodiesterase | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Phosphodiesterase, RETINAL | | Authors: | Ikuta, T, Shihoya, W, Yamashita, K, Nureki, O. | | Deposit date: | 2020-10-05 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural insights into the mechanism of rhodopsin phosphodiesterase.
Nat Commun, 11, 2020
|
|
11AS
 
 | |
12AS
 
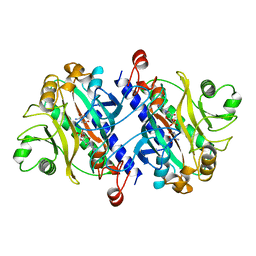 | | ASPARAGINE SYNTHETASE MUTANT C51A, C315A COMPLEXED WITH L-ASPARAGINE AND AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ASPARAGINE, ASPARAGINE SYNTHETASE | | Authors: | Nakatsu, T, Kato, H, Oda, J. | | Deposit date: | 1997-12-02 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of asparagine synthetase reveals a close evolutionary relationship to class II aminoacyl-tRNA synthetase.
Nat.Struct.Biol., 5, 1998
|
|
7W9Q
 
 | | Crystal structure of V30M-TTR in complex with naringenin derivative-14 | | Descriptor: | (2~{R})-2-(3-chloranyl-4-oxidanyl-phenyl)-5,7-bis(oxidanyl)-2,3-dihydrochromen-4-one, CALCIUM ION, Transthyretin | | Authors: | Katayama, W, Shimane, A, Nabeshima, Y, Yokoyama, T, Mizuguchi, M. | | Deposit date: | 2021-12-10 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Chlorinated Naringenin Analogues as Potential Inhibitors of Transthyretin Amyloidogenesis.
J.Med.Chem., 65, 2022
|
|
