2QZ4
 
 | | Human paraplegin, AAA domain in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Paraplegin | | Authors: | Karlberg, T, Lehtio, L, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Johansson, I, Kallas, A, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, J, Sagemark, C, Sundstrom, M, Thorsell, A.G, Tresauges, L, Van Den Berg, S, Weigelt, J, Welin, M, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-08-16 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal Structure of the ATPase Domain of the Human AAA+ Protein Paraplegin/SPG7.
Plos One, 4, 2009
|
|
2RF5
 
 | | Crystal structure of human tankyrase 1- catalytic PARP domain | | Descriptor: | GLYCEROL, Tankyrase-1, ZINC ION | | Authors: | Lehtio, L, Karlberg, T, Arrowsmith, C.H, Berglund, H, Busam, R, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Holmberg-Schiavone, L, Johansson, I, Kallas, A, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Thorsell, A.G, Tresaugues, L, van den Berg, S, Welin, M, Weigelt, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-28 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Zinc binding catalytic domain of human tankyrase 1.
J.Mol.Biol., 379, 2008
|
|
2R59
 
 | | Leukotriene A4 hydrolase complexed with inhibitor RB3041 | | Descriptor: | ACETIC ACID, Leukotriene A-4 hydrolase, N-{(2S)-3-[(R)-[(1R)-1-amino-2-phenylethyl](hydroxy)phosphoryl]-2-benzylpropanoyl}-L-phenylalanine, ... | | Authors: | Tholander, F, Haeggstrom, J.Z, Thunnissen, M, Muroya, A, Roques, B.P, Fournie-Zaluski, M.C. | | Deposit date: | 2007-09-03 | | Release date: | 2008-08-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure-based dissection of the active site chemistry of leukotriene a4 hydrolase: implications for m1 aminopeptidases and inhibitor design.
Chem.Biol., 15, 2008
|
|
2R55
 
 | | Human StAR-related lipid transfer protein 5 | | Descriptor: | StAR-related lipid transfer protein 5 | | Authors: | Lehtio, L, Busam, R.D, Arrowsmith, C.H, Berglund, H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Herman, M.D, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Sagemark, J, Sundstrom, M, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Persson, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-03 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Comparative structural analysis of lipid binding START domains.
Plos One, 6, 2011
|
|
3Q71
 
 | | Human parp14 (artd8) - macro domain 2 in complex with adenosine-5-diphosphoribose | | Descriptor: | Poly [ADP-ribose] polymerase 14, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Karlberg, T, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Ekblad, T, Flodin, S, Flores, A, Graslund, S, Kotenyova, T, Kouznetsova, E, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sehic, A, Thorsell, A.G, Tresaugues, L, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recognition of Mono-ADP-Ribosylated ARTD10 Substrates by ARTD8 Macrodomains.
Structure, 21, 2013
|
|
3Q6Z
 
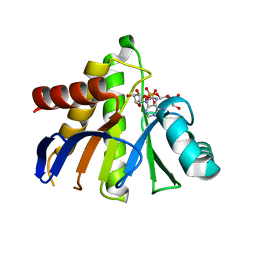 | | HUman PARP14 (ARTD8)-Macro domain 1 in complex with adenosine-5-diphosphoribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Poly [ADP-ribose] polymerase 14 | | Authors: | Karlberg, T, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Ekblad, T, Flodin, S, Flores, A, Graslund, S, Kotenyova, T, Kouznetsova, E, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sehic, A, Thorsell, A.G, Tresaugues, L, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-04 | | Release date: | 2011-02-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Recognition of Mono-ADP-Ribosylated ARTD10 Substrates by ARTD8 Macrodomains.
Structure, 21, 2013
|
|
5MMT
 
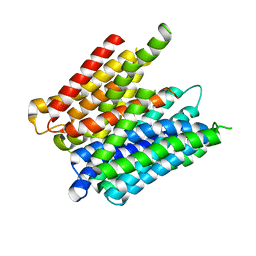 | |
4J1Z
 
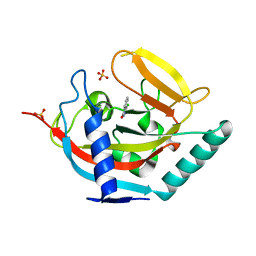 | |
1AV8
 
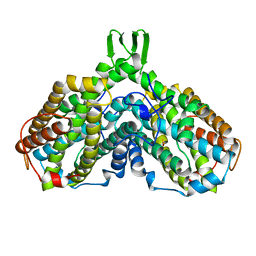 | | RIBONUCLEOTIDE REDUCTASE R2 SUBUNIT FROM E. COLI | | Descriptor: | MU-OXO-DIIRON, RIBONUCLEOTIDE REDUCTASE R2 | | Authors: | Han, S, Arvai, A, Tainer, J.A. | | Deposit date: | 1997-09-30 | | Release date: | 1998-10-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterization of Y122F R2 of Escherichia coli ribonucleotide reductase by time-resolved physical biochemical methods and X-ray crystallography.
Biochemistry, 37, 1998
|
|
2PZ9
 
 | | Crystal structure of putative transcriptional regulator SCO4942 from Streptomyces coelicolor | | Descriptor: | Putative regulatory protein, SULFATE ION | | Authors: | Filippova, E.V, Chruszcz, M, Xu, X, Zheng, H, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-05-17 | | Release date: | 2007-06-19 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | In situ proteolysis for protein crystallization and structure determination.
Nat.Methods, 4, 2007
|
|
3GZC
 
 | | Structure of human selenocysteine lyase | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, Selenocysteine lyase | | Authors: | Collins, R, Hogbom, M, Arrowsmith, C, Berglund, H, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Karlberg, T, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Uppenberg, J, Van Den Berg, S, Weigelt, J, Holmberg-Schiavone, L, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-07 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical discrimination between selenium and sulfur 1: a single residue provides selenium specificity to human selenocysteine lyase.
Plos One, 7, 2012
|
|
3IUY
 
 | | Crystal structure of DDX53 DEAD-box domain | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Probable ATP-dependent RNA helicase DDX53 | | Authors: | Schutz, P, Karlberg, T, Collins, R, Arrowsmith, C.H, Berglund, H, Bountra, C, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kraulis, P, Kotenyova, T, Kotzsch, A, Markova, N, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-31 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Comparative Structural Analysis of Human DEAD-Box RNA Helicases.
Plos One, 5, 2010
|
|
3KJD
 
 | | Human poly(ADP-ribose) polymerase 2, catalytic fragment in complex with an inhibitor ABT-888 | | Descriptor: | (2R)-2-(7-carbamoyl-1H-benzimidazol-2-yl)-2-methylpyrrolidinium, GLYCEROL, Poly [ADP-ribose] polymerase 2 | | Authors: | Karlberg, T, Schutz, P, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kotenyova, T, Kotzsch, A, Kraulis, P, Nielsen, T.K, Moche, M, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-03 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the catalytic domain of human PARP2 in complex with PARP inhibitor ABT-888.
Biochemistry, 49, 2010
|
|
3KCZ
 
 | | Human poly(ADP-ribose) polymerase 2, catalytic fragment in complex with an inhibitor 3-aminobenzamide | | Descriptor: | 3-aminobenzamide, GLYCEROL, Poly [ADP-ribose] polymerase 2 | | Authors: | Karlberg, T, Schutz, P, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kotenyova, T, Kotzsch, A, Kraulis, P, Nielsen, T.K, Moche, M, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-22 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the catalytic domain of human PARP2 in complex with PARP inhibitor ABT-888.
Biochemistry, 49, 2010
|
|
3SE2
 
 | | Human poly(ADP-ribose) polymerase 14 (PARP14/ARTD8) - catalytic domain in complex with 6(5H)-phenanthridinone | | Descriptor: | 3-aminobenzamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Karlberg, T, Schutz, P, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Ekblad, T, Graslund, S, Kouznetsova, E, Moche, M, Nordlund, P, Nyman, T, Thorsell, A.G, Tresaugues, L, Weigelt, J, Siponen, M.I, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-10 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors.
Nat.Biotechnol., 30, 2012
|
|
3GZD
 
 | | Human selenocysteine lyase, P1 crystal form | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, NITRATE ION, Selenocysteine lyase | | Authors: | Karlberg, T, Hogbom, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Schutz, P, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-07 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical discrimination between selenium and sulfur 1: a single residue provides selenium specificity to human selenocysteine lyase.
Plos One, 7, 2012
|
|
3GZF
 
 | | Structure of the C-terminal domain of nsp4 from Feline Coronavirus | | Descriptor: | Replicase polyprotein 1ab, SULFATE ION | | Authors: | Manolaridis, I, Wojdyla, J.A, Panjikar, S, Snijder, E.J, Gorbalenya, A.E, Coutard, B, Tucker, P.A. | | Deposit date: | 2009-04-07 | | Release date: | 2009-08-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.756 Å) | | Cite: | Structure of the C-terminal domain of nsp4 from feline coronavirus
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3HND
 
 | | Crystal structure of human ribonucleotide reductase 1 bound to the effector TTP and substrate GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large subunit, ... | | Authors: | Fairman, J.W, Wijerathna, S.R, Xu, H, Dealwis, C.G. | | Deposit date: | 2009-05-31 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3HNC
 
 | | Crystal structure of human ribonucleotide reductase 1 bound to the effector TTP | | Descriptor: | MAGNESIUM ION, Ribonucleoside-diphosphate reductase large subunit, SULFATE ION, ... | | Authors: | Fairman, J.W, Wijerathna, S.R, Xu, H, Dealwis, C.G. | | Deposit date: | 2009-05-31 | | Release date: | 2011-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3HNE
 
 | | Crystal structure of human ribonucleotide reductase 1 bound to the effectors TTP and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large subunit, ... | | Authors: | Fairman, J.W, Wijerathna, S.R, Xu, H, Dealwis, C.G. | | Deposit date: | 2009-05-31 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3HNF
 
 | | Crystal structure of human ribonucleotide reductase 1 bound to the effectors TTP and dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large subunit, ... | | Authors: | Fairman, J.W, Wijerathna, S.R, Xu, H, Dealwis, C.G. | | Deposit date: | 2009-05-31 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3LLM
 
 | | Crystal Structure Analysis of a RNA Helicase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase A, CACODYLATE ION, ... | | Authors: | Schutz, P, Karlberg, T, Collins, R, Arrowsmith, C.H, Berglund, H, Bountra, C, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kraulis, P, Kotenyova, T, Kotzsch, A, Markova, N, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-29 | | Release date: | 2010-05-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human RNA helicase A (DHX9): structural basis for unselective nucleotide base binding in a DEAD-box variant protein.
J.Mol.Biol., 400, 2010
|
|
3H9U
 
 | | S-adenosyl homocysteine hydrolase (SAHH) from Trypanosoma brucei | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, GLYCEROL, ... | | Authors: | Siponen, M.I, Schutz, P, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kragh Nielsen, T, Kotenyova, T, Kotzsch, A, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schueler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-30 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of S-adenosyl homocysteine hydrolase (SAHH) from Trypanosoma brucei
To be Published
|
|
3IUC
 
 | | Crystal structure of the human 70kDa heat shock protein 5 (BiP/GRP78) ATPase domain in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Heat shock 70kDa protein 5 (Glucose-regulated protein, ... | | Authors: | Wisniewska, M, Karlberg, T, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kotyenova, T, Kotzch, A, Kraulis, P, Markova, N, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A, Schutz, P, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-31 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the ATPase domains of four human Hsp70 isoforms: HSPA1L/Hsp70-hom, HSPA2/Hsp70-2, HSPA6/Hsp70B', and HSPA5/BiP/GRP78
Plos One, 5, 2010
|
|
3JXU
 
 | | Crystal structure of the human 70kDa heat shock protein 1A (Hsp70-1) ATPase domain in complex with ADP and inorganic phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat shock 70 kDa protein 1, MAGNESIUM ION, ... | | Authors: | Wisniewska, M.M, Karlberg, T, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kraulis, P, Kotenyova, T, Kotzsch, A, Markova, N, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Siponen, M.I, Schutz, P, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-21 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structures of the ATPase domains of four human Hsp70 isoforms: HSPA1L/Hsp70-hom, HSPA2/Hsp70-2, HSPA6/Hsp70B', and HSPA5/BiP/GRP78
Plos One, 5, 2010
|
|
