5L9Z
 
 | | Crystal structure of human heparanase nucleophile mutant (E343Q), in complex with unreacted glucuronic acid configured aziridine probe JJB355 | | Descriptor: | (1~{R},2~{S},3~{R},4~{S},5~{S},6~{R})-7-[8-[(azanylidene-{4}-azanylidene)amino]octyl]-3,4,5-tris(oxidanyl)-7-azabicyclo[4.1.0]heptane-2-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, L, Jin, Y, Davies, G.J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Activity-based probes for functional interrogation of retaining beta-glucuronidases.
Nat. Chem. Biol., 13, 2017
|
|
5JAW
 
 | | Structure of a beta galactosidase with inhibitor | | Descriptor: | (1S,2S,3S,4S,5R,6R)-5-amino-6-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, ACETATE ION, Beta-galactosidase, ... | | Authors: | Offen, W, Davies, G. | | Deposit date: | 2016-04-12 | | Release date: | 2017-05-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Towards broad spectrum activity-based glycosidase probes: synthesis and evaluation of deoxygenated cyclophellitol aziridines.
Chem. Commun. (Camb.), 53, 2017
|
|
6B9K
 
 | |
6CI5
 
 | | Crystal structure of the formyltransferase PseJ from Anoxybacillus kamchatkensis in complex with UDP-4,6-dideoxy-4-formamido-L-AltNAc and tetrahydrofolate | | Descriptor: | (2R,3R,4S,5R,6S)-3-(acetylamino)-5-(formylamino)-4-hydroxy-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name), N-[4-({[(6R)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, SULFATE ION, ... | | Authors: | Reimer, J.M, Harb, I, Schmeing, T.M. | | Deposit date: | 2018-02-23 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.00003052 Å) | | Cite: | Structural Insight into a Novel Formyltransferase and Evolution to a Nonribosomal Peptide Synthetase Tailoring Domain.
ACS Chem. Biol., 13, 2018
|
|
3T0T
 
 | | Crystal structure of S. aureus Pyruvate Kinase | | Descriptor: | N'-[(1E)-1-(1H-benzimidazol-2-yl)ethylidene]-5-bromo-2-hydroxybenzohydrazide, PHOSPHATE ION, Pyruvate kinase | | Authors: | Worrall, L.J, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2011-07-20 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Cheminformatics-driven discovery of selective, nanomolar inhibitors for staphylococcal pyruvate kinase.
Acs Chem.Biol., 7, 2012
|
|
6CI4
 
 | | Crystal structure of the formyltransferase PseJ from Anoxybacillus kamchatkensis soaked with UDP-4-amino-4,6-dideoxy-L-AltNAc | | Descriptor: | (2R,3R,4S,5R,6S)-3-(acetylamino)-5-amino-4-hydroxy-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, formyltransferase PseJ | | Authors: | Harb, I, Reimer, J.M, Schmeing, T.M. | | Deposit date: | 2018-02-23 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.824068 Å) | | Cite: | Structural Insight into a Novel Formyltransferase and Evolution to a Nonribosomal Peptide Synthetase Tailoring Domain.
ACS Chem. Biol., 13, 2018
|
|
6BK8
 
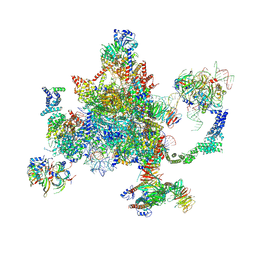 | | S. cerevisiae spliceosomal post-catalytic P complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Lea1, ... | | Authors: | Liu, S, Li, X, Zhou, Z.H, Zhao, R. | | Deposit date: | 2017-11-07 | | Release date: | 2018-02-21 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the yeast spliceosomal postcatalytic P complex.
Science, 358, 2017
|
|
7L4C
 
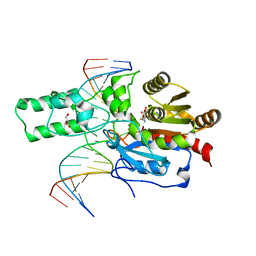 | | Crystal structure of the DRM2-CTT DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*AP*TP*TP*AP*AP*TP*(C49)P*TP*TP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*AP*AP*GP*AP*TP*TP*AP*AP*TP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Fang, J, Song, J. | | Deposit date: | 2020-12-18 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4K
 
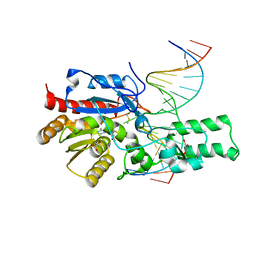 | | Crystal structure of the DRM2-CCG DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*CP*CP*TP*AP*AP*TP*(C49)P*CP*GP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*CP*GP*GP*AP*TP*TP*AP*GP*GP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Fang, J, Song, J. | | Deposit date: | 2020-12-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4H
 
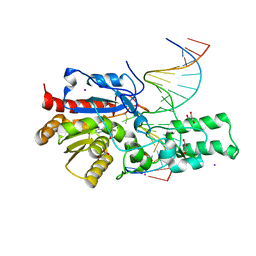 | | Crystal structure of the DRM2-CTG DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*CP*CP*TP*AP*AP*TP*(C49)P*TP*GP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*CP*AP*GP*AP*TP*TP*AP*GP*GP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Fang, J, Song, J. | | Deposit date: | 2020-12-19 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4N
 
 | | Crystal structure of the DRM2 (C397R)-CCG DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*CP*CP*TP*AP*AP*TP*(C49)P*CP*GP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*CP*GP*GP*AP*TP*TP*AP*GP*GP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Fang, J, Song, J. | | Deposit date: | 2020-12-19 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4M
 
 | | Crystal structure of the DRM2-CCT DNA complex | | Descriptor: | DNA (5'-D(*TP*AP*AP*AP*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*GP*AP*AP*T)-3'), DNA (5'-D(P*AP*TP*TP*CP*CP*TP*CP*CP*TP*(C49)P*CP*TP*CP*CP*TP*TP*TP*A)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Fang, J, Song, J. | | Deposit date: | 2020-12-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4F
 
 | | Crystal structure of the DRM2-CAT DNA complex | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, DNA (5'-D(*AP*TP*TP*CP*CP*TP*CP*CP*TP*(C49)P*AP*TP*CP*CP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*GP*GP*AP*TP*GP*AP*GP*GP*AP*GP*GP*AP*AP*T)-3'), ... | | Authors: | Fang, J, Song, J. | | Deposit date: | 2020-12-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
3HA8
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/Compound 14b | | Descriptor: | Mitogen-activated protein kinase 14, N~2~-{4-[6-(3,4-dihydroquinolin-1(2H)-ylcarbonyl)-1H-benzimidazol-1-yl]-6-ethoxy-1,3,5-triazin-2-yl}-3-(2,2-dimethyl-4H-1,3-benzodioxin-6-yl)-N-methyl-L-alaninamide | | Authors: | Zhao, B, Clark, M.A. | | Deposit date: | 2009-05-01 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Design, synthesis and selection of DNA-encoded small-molecule libraries.
Nat.Chem.Biol., 5, 2009
|
|
3HA6
 
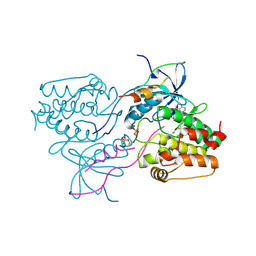 | | Crystal structure of aurora A in complex with TPX2 and compound 10 | | Descriptor: | N~2~-(3,4-dimethoxyphenyl)-N~4~-[2-(2-fluorophenyl)ethyl]-N~6~-quinolin-6-yl-1,3,5-triazine-2,4,6-triamine, Serine/threonine-protein kinase 6, Targeting protein for Xklp2 | | Authors: | Zhao, B, Clark, M.A. | | Deposit date: | 2009-05-01 | | Release date: | 2009-08-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Design, synthesis and selection of DNA-encoded small-molecule libraries.
Nat.Chem.Biol., 5, 2009
|
|
5G0Q
 
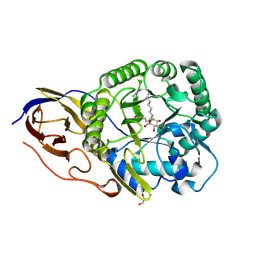 | | beta-glucuronidase with an activity-based probe (N-alkyl cyclophellitol aziridine) bound | | Descriptor: | (1R,2S,3R,4S,5S,6R)-7-(8-AZIDOOCTYL)-3,4,5-TRIHYDROXY-, BETA-GLUCURONIDASE, GLYCEROL | | Authors: | Jin, Y, Wu, L, Jiang, J.B, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2016-03-22 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Activity-based probes for functional interrogation of retaining beta-glucuronidases.
Nat. Chem. Biol., 13, 2017
|
|
4QLC
 
 | | Crystal structure of chromatosome at 3.5 angstrom resolution | | Descriptor: | CITRIC ACID, DNA (167-mer), H5, ... | | Authors: | Jiang, J.S, Zhou, B.R, Xiao, T.S, Bai, Y.W. | | Deposit date: | 2014-06-11 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Structural Mechanisms of Nucleosome Recognition by Linker Histones.
Mol.Cell, 33 Suppl 1, 2015
|
|
3VD8
 
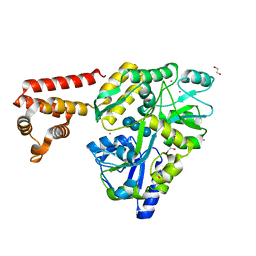 | | Crystal structure of human AIM2 PYD domain with MBP fusion | | Descriptor: | 1,2-ETHANEDIOL, Maltose-binding periplasmic protein, Interferon-inducible protein AIM2, ... | | Authors: | Jin, T.C, Perry, A, Smith, P, Xiao, T.S. | | Deposit date: | 2012-01-04 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.0685 Å) | | Cite: | Structure of the Absent in Melanoma 2 (AIM2) Pyrin Domain Provides Insights into the Mechanisms of AIM2 Autoinhibition and Inflammasome Assembly.
J.Biol.Chem., 288, 2013
|
|
7XR4
 
 | |
7XR6
 
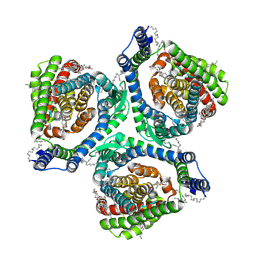 | | Structure of human excitatory amino acid transporter 2 (EAAT2) in complex with WAY-213613 | | Descriptor: | (2S)-2-azanyl-4-[[4-[2-bromanyl-4,5-bis(fluoranyl)phenoxy]phenyl]amino]-4-oxidanylidene-butanoic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Zhao, Y, Zhang, Z. | | Deposit date: | 2022-05-09 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of ligand binding modes of human EAAT2.
Nat Commun, 13, 2022
|
|
7LC9
 
 | |
4KMD
 
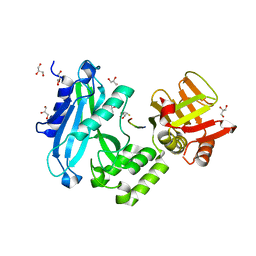 | | Crystal structure of Sufud60-Gli1p | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, Sufu, ... | | Authors: | Zhang, Y, Qi, X, Zhang, Z, Wu, G. | | Deposit date: | 2013-05-08 | | Release date: | 2013-11-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into the mutual recognition and regulation between Suppressor of Fused and Gli/Ci.
Nat Commun, 4, 2013
|
|
5FNV
 
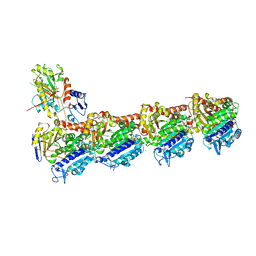 | | a new complex structure of tubulin with an alpha-beta unsaturated lactone | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, Y, Naismith, J, Zhu, X. | | Deposit date: | 2015-11-16 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Pironetin Reacts Covalently with Cysteine-316 of Alpha-Tubulin to Destabilize Microtubule.
Nat.Commun., 7, 2016
|
|
4KMH
 
 | |
4KM9
 
 | |
