4TTH
 
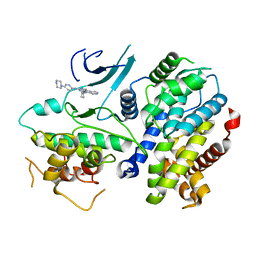 | | Crystal structure of a CDK6/Vcyclin complex with inhibitor bound | | Descriptor: | 9-cyclopentyl-N-(5-piperazin-1-ylpyridin-2-yl)pyrido[4,5]pyrrolo[1,2-d]pyrimidin-2-amine, Cyclin homolog, Cyclin-dependent kinase 6 | | Authors: | Piper, D.E, Walker, N, Wang, Z. | | Deposit date: | 2014-06-20 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of AMG 925, a FLT3 and CDK4 dual kinase inhibitor with preferential affinity for the activated state of FLT3.
J.Med.Chem., 57, 2014
|
|
4R8W
 
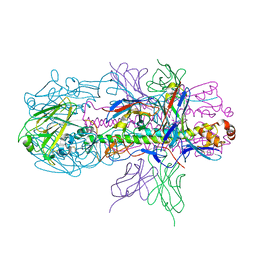 | | Crystal structure of H7 hemagglutinin from A/Anhui/1/2013 in complex with a neutralizing antibody CT149 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of neutralizing antibody CT149, Hemagglutinin, ... | | Authors: | Wu, Y, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2014-09-03 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.795 Å) | | Cite: | A potent broad-spectrum protective human monoclonal antibody crosslinking two haemagglutinin monomers of influenza A virus
Nat Commun, 6, 2015
|
|
4ZJV
 
 | |
7C0J
 
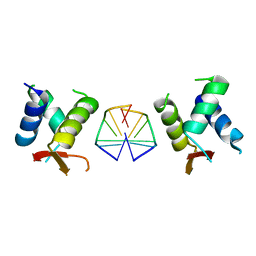 | | Crystal structure of chimeric mutant of GH5 in complex with Z-DNA | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*CP*G)-3'), Histone H5,Double-stranded RNA-specific adenosine deaminase | | Authors: | Choi, H.J, Park, C.H. | | Deposit date: | 2020-05-01 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Dual conformational recognition by Z-DNA binding protein is important for the B-Z transition process.
Nucleic Acids Res., 48, 2020
|
|
7C0I
 
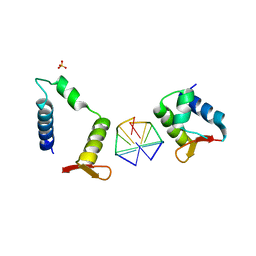 | | Crystal structure of chimeric mutant of E3L in complex with Z-DNA | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*CP*G)-3'), Double-stranded RNA-binding protein,Double-stranded RNA-specific adenosine deaminase, SULFATE ION | | Authors: | Choi, H.J, Park, C.H, Kim, J.S. | | Deposit date: | 2020-05-01 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dual conformational recognition by Z-DNA binding protein is important for the B-Z transition process.
Nucleic Acids Res., 48, 2020
|
|
2GGR
 
 | |
6W54
 
 | | Crystal Structure of Gallic Acid Decarboxylase from Arxula adeninivorans | | Descriptor: | 4-NITROCATECHOL, COBALT (II) ION, Gallate decarboxylase, ... | | Authors: | Zeug, M, Marckovic, N, Iancu, C.V, Tripp, J, Oreb, M, Choe, J. | | Deposit date: | 2020-03-12 | | Release date: | 2021-02-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of non-oxidative decarboxylases reveal a new mechanism of action with a catalytic dyad and structural twists.
Sci Rep, 11, 2021
|
|
2Q7D
 
 | | Crystal Structure of Human Inositol 1,3,4-Trisphosphate 5/6-kinase (ITPK1) in complex with AMPPNP and Mn2+ | | Descriptor: | Inositol-tetrakisphosphate 1-kinase, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chamberlain, P.P, Lesley, S.A, Spraggon, G. | | Deposit date: | 2007-06-06 | | Release date: | 2007-07-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Integration of inositol phosphate signaling pathways via human ITPK1.
J.Biol.Chem., 282, 2007
|
|
2QB5
 
 | | Crystal Structure of Human Inositol 1,3,4-Trisphosphate 5/6-Kinase (ITPK1) in Complex with ADP and Mn2+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Inositol-tetrakisphosphate 1-kinase, MANGANESE (II) ION, ... | | Authors: | Chamberlain, P.P, Lesley, S.A, Spraggon, G. | | Deposit date: | 2007-06-15 | | Release date: | 2007-07-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Integration of inositol phosphate signaling pathways via human ITPK1.
J.Biol.Chem., 282, 2007
|
|
4KDL
 
 | | Crystal structure of p97/VCP N in complex with OTU1 UBXL | | Descriptor: | Transitional endoplasmic reticulum ATPase, Ubiquitin thioesterase OTU1 | | Authors: | Kim, S.J, Kim, E.E. | | Deposit date: | 2013-04-25 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Basis for Ovarian Tumor Domain-containing Protein 1 (OTU1) Binding to p97/Valosin-containing Protein (VCP).
J.Biol.Chem., 289, 2014
|
|
4ONJ
 
 | | Crystal structure of the catalytic domain of ntDRM | | Descriptor: | DNA methyltransferase, SINEFUNGIN | | Authors: | Du, J, Patel, D.J. | | Deposit date: | 2014-01-28 | | Release date: | 2014-06-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | Molecular Mechanism of Action of Plant DRM De Novo DNA Methyltransferases.
Cell(Cambridge,Mass.), 157, 2014
|
|
4ONQ
 
 | |
6Q9L
 
 | |
6Q9U
 
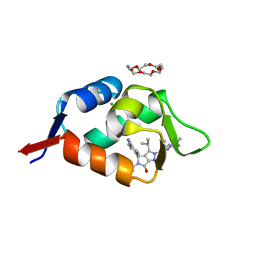 | | HDMX (14-111; C17S) COMPLEXED WITH COMPOUND 12 AT 2.4A; Structural states of Hdm2 and HdmX: X-ray elucidation of adaptations and binding interactions for different chemical compound classes | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 4-[(4~{S})-5-(5-chloranyl-2-oxidanylidene-1~{H}-pyridin-3-yl)-2-[2-(dimethylamino)-4-methoxy-pyrimidin-5-yl]-6-oxidanylidene-3-propan-2-yl-4~{H}-pyrrolo[3,4-c]pyrazol-4-yl]benzenecarbonitrile, CHLORIDE ION, ... | | Authors: | Kallen, J. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural States of Hdm2 and HdmX: X-ray Elucidation of Adaptations and Binding Interactions for Different Chemical Compound Classes.
Chemmedchem, 14, 2019
|
|
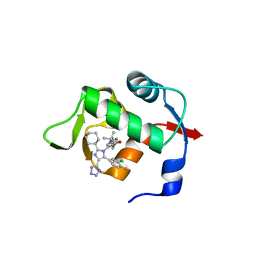
6Q9O
 
 | |
6Q9Q
 
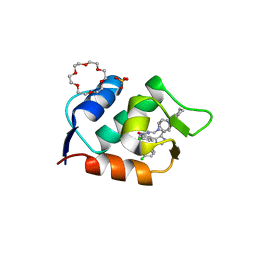 | | HDMX (14-111; C17S) COMPLEXED WITH COMPOUND 13 AT 2.1A; Structural states of Hdm2 and HdmX: X-ray elucidation of adaptations and binding interactions for different chemical compound classes | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 6-chloranyl-3-[3-[(1~{S})-1-(4-chlorophenyl)ethyl]-5-phenyl-imidazol-4-yl]-~{N}-[2-(4-cyclohexylpiperazin-1-yl)ethyl]-1~{H}-indole-2-carboxamide, GLYCEROL, ... | | Authors: | Kallen, J. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural States of Hdm2 and HdmX: X-ray Elucidation of Adaptations and Binding Interactions for Different Chemical Compound Classes.
Chemmedchem, 14, 2019
|
|
6Q9W
 
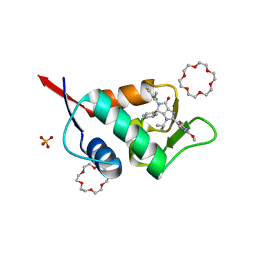 | | X-ray structure of compound 15 bound to HdmX: Structural states of Hdm2 and HdmX: X-ray elucidation of adaptations and binding interactions for different chemical compound classes | | Descriptor: | (4~{S})-4-(4-chlorophenyl)-5-[(1~{S})-1-(3-chlorophenyl)ethyl]-2-(2,4-dimethoxypyrimidin-5-yl)-3-propan-2-yl-4~{H}-pyrrolo[3,4-d]imidazol-6-one, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Protein Mdm4, ... | | Authors: | Kallen, J. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural States of Hdm2 and HdmX: X-ray Elucidation of Adaptations and Binding Interactions for Different Chemical Compound Classes.
Chemmedchem, 14, 2019
|
|
6Q9H
 
 | |
6Q9Y
 
 | |
6Q9S
 
 | | HDMX (14-111; C17S) COMPLEXED WITH COMPOUND 14 AT 2.4A: Structural states of Hdm2 and HdmX: X-ray elucidation of adaptations and binding interactions for different chemical compound classes | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 3-[[6-chloranyl-3-[3-[(1~{S})-1-(2,4-dichlorophenyl)ethyl]-5-phenyl-imidazol-4-yl]-1~{H}-indol-2-yl]carbonylamino]-4-[4-(2-oxidanylidene-1,3-oxazinan-3-yl)piperidin-1-yl]benzoic acid, Protein Mdm4, ... | | Authors: | Kallen, J. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural States of Hdm2 and HdmX: X-ray Elucidation of Adaptations and Binding Interactions for Different Chemical Compound Classes.
Chemmedchem, 14, 2019
|
|
6Q96
 
 | |
7BOL
 
 | | ubiquitin-conjugating enzyme, Ube2D2 | | Descriptor: | Ubiquitin-conjugating enzyme E2 D2 | | Authors: | Lee, S.O, Ryu, K.S, Chi, S.-W. | | Deposit date: | 2020-03-19 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | MUL1-RING recruits the substrate, p53-TAD as a complex with UBE2D2-UB conjugate.
Febs J., 2022
|
|
1D4B
 
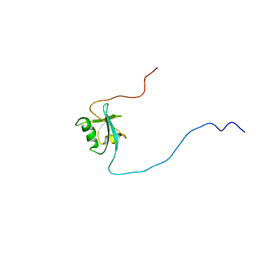 | | CIDE-N DOMAIN OF HUMAN CIDE-B | | Descriptor: | HUMAN CELL DEATH-INDUCING EFFECTOR B | | Authors: | Lugovskoy, A, Zhou, P, Chou, J, McCarty, J, Li, P, Wagner, G. | | Deposit date: | 1999-10-02 | | Release date: | 1999-12-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CIDE-N domain of CIDE-B and a model for CIDE-N/CIDE-N interactions in the DNA fragmentation pathway of apoptosis.
Cell(Cambridge,Mass.), 99, 1999
|
|
1CJ1
 
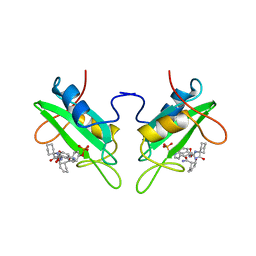 | | GROWTH FACTOR RECEPTOR BINDING PROTEIN SH2 DOMAIN (HUMAN) COMPLEXED WITH A PHOSPHOTYROSYL DERIVATIVE | | Descriptor: | PROTEIN (GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2), [1-[1-(6-CARBAMOYL-CYCLOHEX-2-ENYLCARBAMOYL)-CYCLOHEXYLCARBAMOYL]-2-(4-PHOSPHONOOXY-PHENYL)- ETHYL]-CARBAMIC ACID 3-AMINOBENZYLESTER | | Authors: | Rahuel, J. | | Deposit date: | 1999-04-21 | | Release date: | 1999-12-22 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based design, synthesis, and X-ray crystallography of a high-affinity antagonist of the Grb2-SH2 domain containing an asparagine mimetic.
J.Med.Chem., 42, 1999
|
|
7L0N
 
 | | Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Snell, G, Czudnochowski, N, Dillen, J, Nix, J.C, Croll, T.I, Corti, D. | | Deposit date: | 2020-12-11 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity.
Cell, 184, 2021
|
|
