7WCN
 
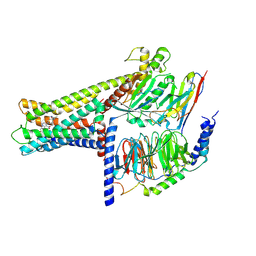 | | Cryo-EM structure of GPR119-Gs Complex with small molecule agonist AR231453 | | Descriptor: | Glucose-dependent insulinotropic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qiao, A.N, Wu, S, Ye, S. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Activation and signaling mechanism revealed by GPR119-G s complex structures.
Nat Commun, 13, 2022
|
|
7VKV
 
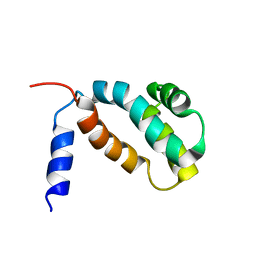 | |
8F5Y
 
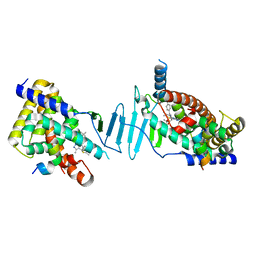 | | Crystal structure of pregnane X receptor ligand binding domain complexed with JQ1 | | Descriptor: | (6S)-6-(2-tert-butoxy-2-oxoethyl)-4-(4-chlorophenyl)-2,3,9-trimethyl-6,7-dihydrothieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-10-ium, Nuclear receptor coactivator 1, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Huber, A.D, Poudel, S, Seetharaman, J, Miller, D.J, Chen, T. | | Deposit date: | 2022-11-15 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A bromodomain-independent mechanism of gene regulation by the BET inhibitor JQ1: direct activation of nuclear receptor PXR.
Nucleic Acids Res., 52, 2024
|
|
7EWQ
 
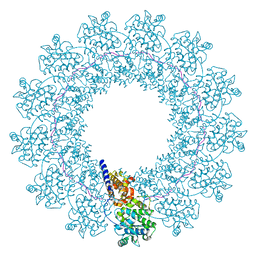 | | Structure of Mumps virus nucleocapsid ring | | Descriptor: | Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Su, X, Shen, Q, Shan, H. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-23 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural plasticity of mumps virus nucleocapsids with cryo-EM structures.
Commun Biol, 4, 2021
|
|
7EXA
 
 | | Structure of mumps virus nucleoprotein without C-arm | | Descriptor: | Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Shen, Q, Shan, H, Zhang, N, Qin, Y. | | Deposit date: | 2021-05-26 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural plasticity of mumps virus nucleocapsids with cryo-EM structures.
Commun Biol, 4, 2021
|
|
7V1Q
 
 | | Leifsonia Alcohol Dehydrogenases LnADH | | Descriptor: | Alcohol Dehydrogenases, ISOPROPYL ALCOHOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Song, Y, Qu, X. | | Deposit date: | 2021-08-05 | | Release date: | 2021-12-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Engineering Leifsonia Alcohol Dehydrogenase for Thermostability and Catalytic Efficiency by Enhancing Subunit Interactions.
Chembiochem, 22, 2021
|
|
7V1R
 
 | | Leifsonia Alcohol Dehydrogenases LnADH | | Descriptor: | Alcohol Dehydrogenases, ISOPROPYL ALCOHOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Song, Y, Qu, X. | | Deposit date: | 2021-08-05 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Engineering Leifsonia Alcohol Dehydrogenase for Thermostability and Catalytic Efficiency by Enhancing Subunit Interactions.
Chembiochem, 22, 2021
|
|
8T4S
 
 | |
6BG5
 
 | | Structure of 1-(benzo[d][1,3]dioxol-5-ylmethyl)-1-(1-propylpiperidin-4-yl)-3-(3-(trifluoromethyl)phenyl)urea bound to DCN1 | | Descriptor: | Endolysin, DCN1-like protein 1 chimera, N-[(2H-1,3-benzodioxol-5-yl)methyl]-N-(1-propylpiperidin-4-yl)-N'-[3-(trifluoromethyl)phenyl]urea | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-10-27 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Piperidinyl Ureas Chemically Control Defective in Cullin Neddylation 1 (DCN1)-Mediated Cullin Neddylation.
J. Med. Chem., 61, 2018
|
|
6BG3
 
 | | Structure of (3S,4S)-1-benzyl-4-(3-(3-(trifluoromethyl)phenyl)ureido)piperidin-3-yl acetate bound to DCN1 | | Descriptor: | Endolysin, DCN1-like protein 1 chimera, N-{(3S,4S)-1-benzyl-3-[(1S)-1-hydroxyethoxy]piperidin-4-yl}-N'-[3-(trifluoromethyl)phenyl]urea | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-10-27 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Piperidinyl Ureas Chemically Control Defective in Cullin Neddylation 1 (DCN1)-Mediated Cullin Neddylation.
J. Med. Chem., 61, 2018
|
|
4NQF
 
 | |
8UVR
 
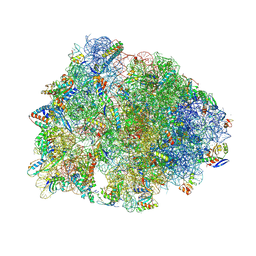 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with spectinomycin, mRNA, deacylated A- and E-site tRNAphe, and deacylated P-site tRNAmet at 2.60A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Killam, B.Y, Phelps, G.A, Lee, R.E, Polikanov, Y.S. | | Deposit date: | 2023-11-03 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Development of 2nd generation aminomethyl spectinomycins that overcome native efflux in Mycobacterium abscessus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UVS
 
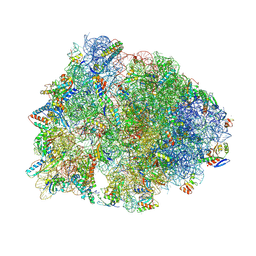 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with spectinomycin derivative 2694, mRNA, deacylated A- and E-site tRNAphe, and deacylated P-site tRNAmet at 2.75A resolution | | Descriptor: | (2R,4R,4aS,5aR,6S,7S,8R,9S,9aR,10aS)-2-methyl-6,8-bis(methylamino)-4-({[2-(oxan-4-yl)ethyl]amino}methyl)octahydro-2H-pyrano[2,3-b][1,4]benzodioxine-4,4a,7,9(10aH)-tetrol, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Killam, B.Y, Phelps, G.A, Lee, R.E, Polikanov, Y.S. | | Deposit date: | 2023-11-03 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Development of 2nd generation aminomethyl spectinomycins that overcome native efflux in Mycobacterium abscessus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5XS4
 
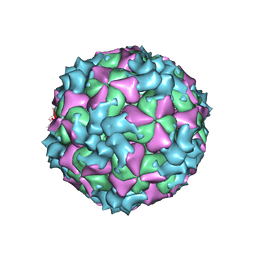 | | Structure of Coxsackievirus A6 (CVA6) virus A-particle | | Descriptor: | Genome polyprotein | | Authors: | Zheng, Q.B, He, M.Z, Xu, L.F, Yu, H, Li, S.W, Cheng, T. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody
Nat Commun, 8, 2017
|
|
7JN5
 
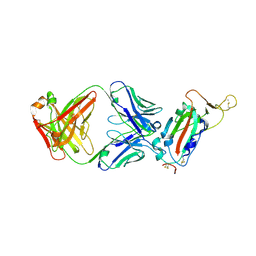 | | Crystal structure of SARS-CoV receptor binding domain in complex with human antibody CR3022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CR3022 heavy chain, ... | | Authors: | Wu, N.C, Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-08-03 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | A natural mutation between SARS-CoV-2 and SARS-CoV determines neutralization by a cross-reactive antibody.
Plos Pathog., 16, 2020
|
|
7JJG
 
 | |
5XS7
 
 | | Structure of Coxsackievirus A6 (CVA6) virus A-particle in complex with the neutralizing antibody fragment 1D5 | | Descriptor: | Genome polyprotein, Heavy chain of Fab 1D5, Light chain of Fab 1D5 | | Authors: | Zheng, Q.B, He, M.Z, Xu, L.F, Yu, H, Li, S.W, Cheng, T. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody
Nat Commun, 8, 2017
|
|
7JJH
 
 | |
7JSP
 
 | | Crystal structure of the second bromodomain (BD2) of human TAF1 bound to ATR kinase inhibitor AZD6738 | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-{1-[(R)-amino(hydroxy)methyl-lambda~4~-sulfanyl]cyclopropyl}-6-[(3R)-3-methylmorpholin-4-yl]pyrimidin-2-yl)-1H-pyrrolo[2,3-b]pyridine, SULFATE ION, ... | | Authors: | Karim, M.R, Schonbrunn, E. | | Deposit date: | 2020-08-15 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Dual TAF1-ATR Inhibitors and Ligand-Induced Structural Changes of the TAF1 Tandem Bromodomain.
J.Med.Chem., 65, 2022
|
|
7K0D
 
 | |
7K0U
 
 | |
7K03
 
 | |
7K42
 
 | | Crystal structure of the second bromodomain (BD2) of human TAF1 bound to dioxane | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, SULFATE ION, ... | | Authors: | Karim, M.R, Schonbrunn, E. | | Deposit date: | 2020-09-14 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Dual TAF1-ATR Inhibitors and Ligand-Induced Structural Changes of the TAF1 Tandem Bromodomain.
J.Med.Chem., 65, 2022
|
|
7K1P
 
 | |
7K27
 
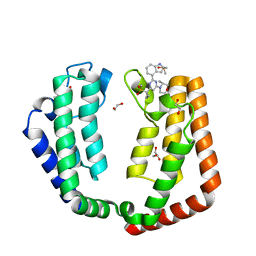 | | Crystal structure of the tandem bromodomain (BD1, BD2) of human TAF1 bound to ATR inhibitor AZ20 | | Descriptor: | 1,2-ETHANEDIOL, 4-{4-[(3R)-3-methylmorpholin-4-yl]-6-[1-(methylsulfonyl)cyclopropyl]pyrimidin-2-yl}-1H-indole, DIMETHYL SULFOXIDE, ... | | Authors: | Karim, M.R, Schonbrunn, E. | | Deposit date: | 2020-09-08 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Dual TAF1-ATR Inhibitors and Ligand-Induced Structural Changes of the TAF1 Tandem Bromodomain.
J.Med.Chem., 65, 2022
|
|
