4X9F
 
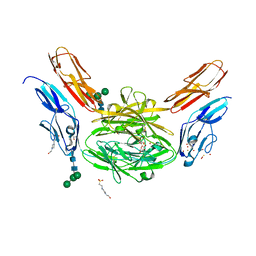 | | Crystal structure of Dscam1 isoform 6.9, N-terminal four Ig domains | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Down Syndrome Cell Adhesion Molecule isoform 6.9, GLYCEROL, ... | | Authors: | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | Deposit date: | 2014-12-11 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4XB7
 
 | | Crystal structure of Dscam1 isoform 4.4, N-terminal four Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Down syndrome cell adhesion molecule, isoform 4.4, ... | | Authors: | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | Deposit date: | 2014-12-16 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4.004 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4XB8
 
 | | Crystal structure of Dscam1 isoform 9.44, N-terminal four Ig domains (with zinc) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Down Syndrome Cell Adhesion Molecule, ... | | Authors: | Chen, Q, Yu, Y, Li, S.A, cheng, L. | | Deposit date: | 2014-12-16 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4X9I
 
 | | Crystal structure of Dscam1 isoform 9.44, N-terminal four Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Down Syndrome Cell Adhesion Molecule, isoform 9.44, ... | | Authors: | Chen, Q, Yu, Y, Li, S.A, cheng, L. | | Deposit date: | 2014-12-11 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.904 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4X5L
 
 | | Crystal structure of Dscam1 Ig7 domain, isoform 9 | | Descriptor: | Down syndrome cell adhesion molecule, isoform AM, SODIUM ION | | Authors: | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | Deposit date: | 2014-12-05 | | Release date: | 2015-12-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.374 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4X9B
 
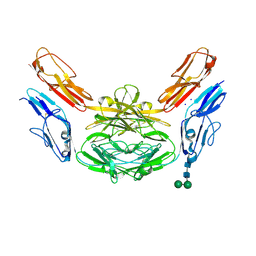 | | Crystal structure of Dscam1 isoform 4.44, N-terminal four Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Down syndrome cell adhesion molecule, isoform 4.44, ... | | Authors: | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | Deposit date: | 2014-12-11 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4X83
 
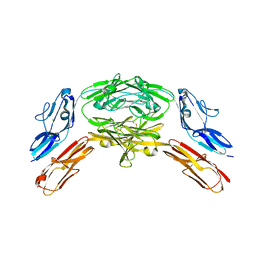 | | Crystal structure of Dscam1 isoform 7.44, N-terminal four Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | Deposit date: | 2014-12-10 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4WVR
 
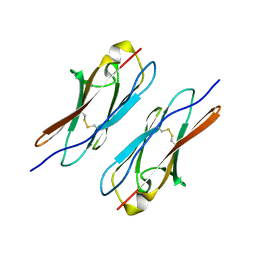 | | Crystal structure of Dscam1 Ig7 domain, isoform 5 | | Descriptor: | Down syndrome cell adhesion molecule, isoform AK | | Authors: | Chen, Q, Yu, Y, Li, S, Cheng, L. | | Deposit date: | 2014-11-07 | | Release date: | 2015-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
9M36
 
 | | Structure of human TRPC5 in the low Ca environment. | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CHOLESTEROL HEMISUCCINATE, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Song, K.C, Wei, M, Chen, L. | | Deposit date: | 2025-02-28 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Structure of human TRPC5 in the low Ca environment.
To Be Published
|
|
5XP7
 
 | | C-Src in complex with ATP-CHCl | | Descriptor: | GLYCEROL, MAGNESIUM ION, Proto-oncogene tyrosine-protein kinase Src, ... | | Authors: | Guo, M, Dai, S, Duan, Y, Chen, L, Chen, Y. | | Deposit date: | 2017-06-01 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Remarkably Stereospecific Utilization of ATP alpha , beta-Halomethylene Analogues by Protein Kinases.
J. Am. Chem. Soc., 139, 2017
|
|
2YR1
 
 | | Crystal Structure of 3-dehydroquinate dehydratase from Geobacillus kaustophilus HTA426 | | Descriptor: | 3-dehydroquinate dehydratase | | Authors: | Kagawa, W, Kurumizaka, H, Bessho, Y, Chen, L, Fu, Z.Q, Chrzas, J, Wang, B.C, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-01 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of 3-dehydroquinate dehydratase from Geobacillus kaustophilus HTA426
To be published
|
|
2YXM
 
 | | Crystal structure of I-set domain of human Myosin Binding ProteinC | | Descriptor: | Myosin-binding protein C, slow-type | | Authors: | Kishishita, S, Ohsawa, N, Murayama, K, Chen, L, Liu, Z, Terada, T, Shirouzu, M, Wang, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-26 | | Release date: | 2007-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of I-set domain of human Myosin Binding ProteinC
To be Published
|
|
2YZ8
 
 | | Crystal structure of the 32th Ig-like domain of human obscurin (KIAA1556) | | Descriptor: | Obscurin | | Authors: | Saijo, S, Ohsawa, N, Nishino, A, Kishishita, S, Chen, L, Fu, Z.Q, Chrzas, J, Wang, B.C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-04 | | Release date: | 2008-05-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the 32th Ig-like domain of human obscurin (KIAA1556)
To be Published
|
|
2YVI
 
 | | Crystal structure of a death domain of human ankryn protein | | Descriptor: | Ankyrin-1, GLYCEROL | | Authors: | Ihsanawati, Bessho, Y, Chen, L, Liu, Z.J, Wang, B.C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-12 | | Release date: | 2008-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of a death domain of human ankryn protein
To be Published
|
|
2YYO
 
 | | Crystal structure of human SPRY domain | | Descriptor: | SPRY domain-containing protein 3 | | Authors: | Kishishita, S, Uchikubo-Kamo, T, Murayama, K, Terada, T, Chen, L, Fu, Z.Q, Chrzas, J, Shirouzu, M, Wang, B.C, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-30 | | Release date: | 2008-05-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human SPRY domain
To be Published
|
|
2Z0M
 
 | | Crystal structure of hypothetical ATP-dependent RNA helicase from Sulfolobus tokodaii | | Descriptor: | 337aa long hypothetical ATP-dependent RNA helicase deaD | | Authors: | Nakagawa, N, Kusano, S, Shirouzu, M, Chen, L, Fu, Z.-Q, Chrzas, J, Wang, B.-C, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of hypothetical ATP-dependent RNA helicase from Sulfolobus tokodaii
To be Published
|
|
4YOM
 
 | | Structure of SAD kinase | | Descriptor: | 1,2-ETHANEDIOL, Serine/threonine-protein kinase BRSK2 | | Authors: | Wu, J.X, Wang, J, Chen, L, Wang, Z.X, Wu, J.W. | | Deposit date: | 2015-03-12 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural insight into the mechanism of synergistic autoinhibition of SAD kinases
Nat Commun, 6, 2015
|
|
4YNZ
 
 | | Structure of the N-terminal domain of SAD | | Descriptor: | Serine/threonine-protein kinase BRSK2 | | Authors: | Wu, J.X, Wang, J, Chen, L, Wang, Z.X, Wu, J.W. | | Deposit date: | 2015-03-11 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into the mechanism of synergistic autoinhibition of SAD kinases
Nat Commun, 6, 2015
|
|
4HQP
 
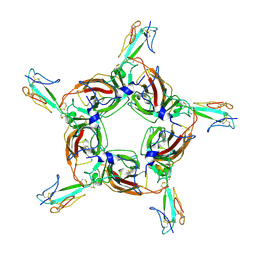 | | Alpha7 nicotinic receptor chimera and its complex with Alpha bungarotoxin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-bungarotoxin isoform V31, ... | | Authors: | Li, S.X, Cheng, K, Gomoto, R, Bren, N, Huang, S, Sine, S, Chen, L. | | Deposit date: | 2012-10-25 | | Release date: | 2013-07-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structural principles for Alpha-neurotoxin binding to and selectivity among nicotinic receptors
To be Published
|
|
3DFX
 
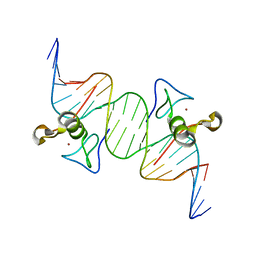 | | Opposite GATA DNA binding | | Descriptor: | DNA (5'-D(*DAP*DAP*DGP*DGP*DTP*DTP*DAP*DTP*DCP*DTP*DCP*DTP*DGP*DAP*DTP*DTP*DTP*DAP*DTP*DC)-3'), DNA (5'-D(*DTP*DTP*DGP*DAP*DTP*DAP*DAP*DAP*DTP*DCP*DAP*DGP*DAP*DGP*DAP*DTP*DAP*DAP*DCP*DC)-3'), Trans-acting T-cell-specific transcription factor GATA-3, ... | | Authors: | Bates, D.L, Kim, G.K, Guo, L, Chen, L. | | Deposit date: | 2008-06-12 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of multiple GATA zinc fingers bound to DNA reveal new insights into DNA recognition and self-association by GATA.
J.Mol.Biol., 381, 2008
|
|
5WUA
 
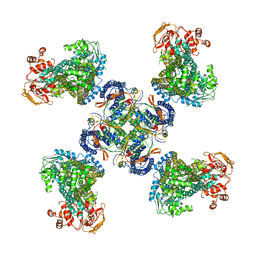 | | Structure of a Pancreatic ATP-sensitive Potassium Channel | | Descriptor: | ATP-sensitive inward rectifier potassium channel 11,superfolder GFP, SUR1 | | Authors: | Li, N, Wu, J.-X, Chen, L, Gao, N. | | Deposit date: | 2016-12-16 | | Release date: | 2017-01-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structure of a Pancreatic ATP-Sensitive Potassium Channel
Cell, 168, 2017
|
|
5X07
 
 | | Crystal structure of FOXA2 DNA binding domain bound to a full consensus DNA site | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*TP*TP*TP*G)-3'), Hepatocyte nuclear factor 3-beta | | Authors: | Li, J, Guo, M, Zhou, Z, Jiang, L, Chen, X, Qu, L, Wu, D, Chen, Z, Chen, L, Chen, Y. | | Deposit date: | 2017-01-20 | | Release date: | 2017-08-16 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Structure of the Forkhead Domain of FOXA2 Bound to a Complete DNA Consensus Site
Biochemistry, 56, 2017
|
|
4WK8
 
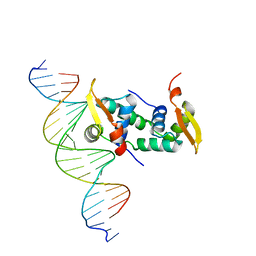 | | FOXP3 forms a domain-swapped dimer to bridge DNA | | Descriptor: | DNA (5'-D(*AP*AP*CP*TP*AP*TP*GP*AP*AP*AP*CP*AP*AP*AP*TP*TP*TP*TP*CP*CP*T)-3'), DNA (5'-D(*TP*TP*AP*GP*GP*AP*AP*AP*AP*TP*TP*TP*GP*TP*TP*TP*CP*AP*TP*AP*G)-3'), Forkhead box protein P3 | | Authors: | Chen, Y, Chen, L. | | Deposit date: | 2014-10-01 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.4006 Å) | | Cite: | DNA binding by FOXP3 domain-swapped dimer suggests mechanisms of long-range chromosomal interactions.
Nucleic Acids Res., 43, 2015
|
|
5X72
 
 | | The crystal Structure PDE delta in complex with (rac)-p9 | | Descriptor: | (2R)-2-(2-fluorophenyl)-3-phenyl-1,2-dihydroquinazolin-4-one, (2S)-2-(2-fluorophenyl)-3-phenyl-1,2-dihydroquinazolin-4-one, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Jiang, Y, Zhuang, C, Chen, L, Wang, R, Wang, F, Sheng, C. | | Deposit date: | 2017-02-23 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Biology-Inspired Discovery of Novel KRAS-PDE delta Inhibitors
J. Med. Chem., 60, 2017
|
|
4TN8
 
 | | Crystal structure of Thermus Thermophilus thioredoxin solved by sulfur SAD using Swiss Light Source data | | Descriptor: | CHLORIDE ION, Thioredoxin | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-06-03 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
