2MVN
 
 | |
1SX3
 
 | | GroEL14-(ATPgammaS)14 | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Chaudhry, C, Horwich, A.L, Brunger, A.T, Adams, P.D. | | Deposit date: | 2004-03-30 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the structural dynamics of the E.coli chaperonin GroEL using translation-libration-screw crystallographic refinement of intermediate states.
J.Mol.Biol., 342, 2004
|
|
7XRI
 
 | | Feruloyl esterase mutant -S106A | | Descriptor: | Cinnamoyl esterase, ethyl (2E)-3-(4-hydroxy-3-methoxyphenyl)prop-2-enoate | | Authors: | Hwang, J.S, Lee, J.H, Do, H, Lee, C.W. | | Deposit date: | 2022-05-10 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Feruloyl Esterase ( La Fae) from Lactobacillus acidophilus : Structural Insights and Functional Characterization for Application in Ferulic Acid Production.
Int J Mol Sci, 24, 2023
|
|
7Y6D
 
 | |
2Q99
 
 | |
5JI2
 
 | | HslU L199Q in HslUV complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU, ATP-dependent protease subunit HslV, ... | | Authors: | Grant, R.A, Sauer, R.T, Schmitz, K.R, Baytshtok, V. | | Deposit date: | 2016-04-21 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.307 Å) | | Cite: | A Structurally Dynamic Region of the HslU Intermediate Domain Controls Protein Degradation and ATP Hydrolysis.
Structure, 24, 2016
|
|
6JQS
 
 | | Structure of Transcription factor, GerE | | Descriptor: | DNA-binding response regulator | | Authors: | Lee, J.H, Lee, C.W. | | Deposit date: | 2019-04-01 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of a transcription factor, GerE (PaGerE), from spore-forming bacterium Paenisporosarcina sp. TG-14.
Biochem.Biophys.Res.Commun., 513, 2019
|
|
5JI3
 
 | | HslUV complex | | Descriptor: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU, ATP-dependent protease subunit HslV | | Authors: | Grant, R.A, Sauer, R.T, Schmitz, K.R, Baytshtok, V. | | Deposit date: | 2016-04-21 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Structurally Dynamic Region of the HslU Intermediate Domain Controls Protein Degradation and ATP Hydrolysis.
Structure, 24, 2016
|
|
7WKS
 
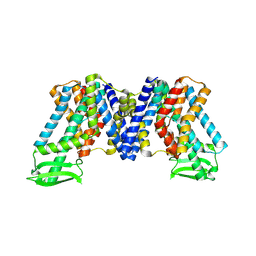 | | Apo state of AtPIN3 | | Descriptor: | Auxin efflux carrier component 3 | | Authors: | Su, N. | | Deposit date: | 2022-01-11 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures and mechanisms of the Arabidopsis auxin transporter PIN3.
Nature, 609, 2022
|
|
7WKW
 
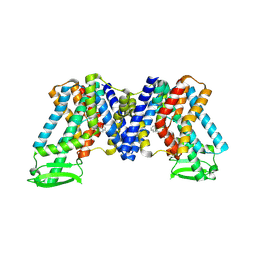 | | NPA bound state of AtPIN3 | | Descriptor: | 2-(naphthalen-1-ylcarbamoyl)benzoic acid, Auxin efflux carrier component 3 | | Authors: | Su, N. | | Deposit date: | 2022-01-11 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structures and mechanisms of the Arabidopsis auxin transporter PIN3.
Nature, 609, 2022
|
|
5YSZ
 
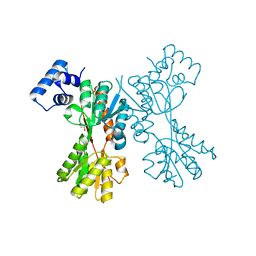 | | transcriptional regulator CelR-cellobiose complex | | Descriptor: | GLYCEROL, Transcriptional regulator, LacI family, ... | | Authors: | Fu, Y, Yeom, S.Y, Lee, D.H, Lee, S.G. | | Deposit date: | 2017-11-16 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Structural and functional analyses of the cellulase transcription regulator CelR
FEBS Lett., 592, 2018
|
|
7XAF
 
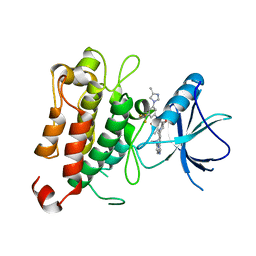 | | The crystal structure of TrkA kinase in complex with 4^6,14-dimethyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-10-oxo-5-oxa-11,14-diaza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclo- tetradecaphan-2-yne-45-carboxamide | | Descriptor: | 4^6,14-dimethyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-10-oxo-5-oxa-11,14-diaza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclo-tetradecaphan-2-yne-45-carboxamide, High affinity nerve growth factor receptor | | Authors: | Zhang, Z.M, Wang, Y.J. | | Deposit date: | 2022-03-17 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.001182 Å) | | Cite: | Discovery of the First Highly Selective and Broadly Effective Macrocycle-Based Type II TRK Inhibitors that Overcome Clinically Acquired Resistance.
J.Med.Chem., 65, 2022
|
|
7X83
 
 | | Cryo-EM structure of the TMEM106B fibril from normal elder | | Descriptor: | Transmembrane protein 106B | | Authors: | Xia, W.C, Zhao, Q.Y, Fan, Y, Sun, Y.P, Tao, Y.Q, Liu, C. | | Deposit date: | 2022-03-11 | | Release date: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Generic amyloid fibrillation of TMEM106B in patient with Parkinson's disease dementia and normal elders.
Cell Res., 32, 2022
|
|
7X84
 
 | | Cryo-EM structure of the TMEM106B fibril from Parkinson's disease dementia | | Descriptor: | Transmembrane protein 106B | | Authors: | Zhao, Q.Y, Xia, W.C, Fan, Y, Sun, Y.P, Tao, Y.Q, Liu, C. | | Deposit date: | 2022-03-11 | | Release date: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Generic amyloid fibrillation of TMEM106B in patient with Parkinson's disease dementia and normal elders.
Cell Res., 32, 2022
|
|
4FYO
 
 | | Crystal structure of spleen tyrosine kinase complexed with N-{(S)-1-[7-(3,4-Dimethoxy-phenylamino)-thiazolo[5,4-d]pyrimidin-5-yl]-pyrrolidin-3-yl}-terephthalamic acid | | Descriptor: | 4-{[(3S)-1-{7-[(3,4-dimethoxyphenyl)amino][1,3]thiazolo[5,4-d]pyrimidin-5-yl}pyrrolidin-3-yl]carbamoyl}benzoic acid, Tyrosine-protein kinase SYK | | Authors: | Kuglstatter, A, Slade, M. | | Deposit date: | 2012-07-05 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rational design of highly selective spleen tyrosine kinase inhibitors.
J.Med.Chem., 55, 2012
|
|
4FZ6
 
 | |
7C4X
 
 | |
8GTZ
 
 | |
4FYN
 
 | |
7ZBV
 
 | |
7Z5U
 
 | | Crystal structure of the peptidase domain of collagenase G from Clostridium histolyticum in complex with a hydroxamate-based inhibitor | | Descriptor: | (2~{R})-~{N}-[2-[4-[(2-acetamidophenoxy)methyl]-1,2,3-triazol-1-yl]ethyl]-2-(2-methylpropyl)-~{N}'-oxidanyl-propanediamide, CALCIUM ION, Collagenase ColG, ... | | Authors: | Schoenauer, E, Brandstetter, H. | | Deposit date: | 2022-03-10 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and Characterization of Synthesized and FDA-Approved Inhibitors of Clostridial and Bacillary Collagenases.
J.Med.Chem., 65, 2022
|
|
7YSE
 
 | | Crystal structure of E. coli heterotetrameric GlyRS in complex with tRNA | | Descriptor: | Glycine--tRNA ligase alpha subunit, Glycine--tRNA ligase beta subunit, MAGNESIUM ION, ... | | Authors: | Han, L, Ju, Y, Zhou, H. | | Deposit date: | 2022-08-12 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.907 Å) | | Cite: | The binding mode of orphan glycyl-tRNA synthetase with tRNA supports the synthetase classification and reveals large domain movements.
Sci Adv, 9, 2023
|
|
7BXZ
 
 | |
7E8N
 
 | | Crystal structure of Type II citrate synthase (HyCS) from Hymenobacter sp. PAMC 26554 | | Descriptor: | CITRIC ACID, Citrate synthase | | Authors: | Park, S.-H, Lee, C.W, Bae, D.-W, Lee, J.H. | | Deposit date: | 2021-03-02 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the cooperative activation of type II citrate synthase (HyCS) from Hymenobacter sp. PAMC 26554.
Int.J.Biol.Macromol., 183, 2021
|
|
7YW3
 
 | | Crystal structure of tRNA 2'-phosphotransferase from Homo sapiens | | Descriptor: | 1,2-ETHANEDIOL, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-3,4-bis(oxidanyl)-5-phosphonooxy-oxolan-2-yl]methyl hydrogen phosphate, tRNA 2'-phosphotransferase 1 | | Authors: | Yang, X.Y, Liu, X.H. | | Deposit date: | 2022-08-21 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical insights into the molecular mechanism of TRPT1 for nucleic acid ADP-ribosylation.
Nucleic Acids Res., 51, 2023
|
|
