184D
 
 | | SELF-ASSOCIATION OF A DNA LOOP CREATES A QUADRUPLEX: CRYSTAL STRUCTURE OF D(GCATGCT) AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*GP*CP*T)-3'), MAGNESIUM ION | | Authors: | Leonard, G.A, Zhang, S, Peterson, M.R, Harrop, S.J, Helliwell, J.R, Cruse, W.B.T, Langlois D'Estaintot, B, Kennard, O, Brown, T, Hunter, W.N. | | Deposit date: | 1994-08-10 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Self-association of a DNA loop creates a quadruplex: crystal structure of d(GCATGCT) at 1.8 A resolution.
Structure, 3, 1995
|
|
7C7Q
 
 | | Cryo-EM structure of the baclofen/BHFF-bound human GABA(B) receptor in active state | | Descriptor: | (3S)-5,7-ditert-butyl-3-oxidanyl-3-(trifluoromethyl)-1-benzofuran-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Mao, C, Shen, C, Li, C, Shen, D, Xu, C, Zhang, S, Zhou, R, Shen, Q, Chen, L, Jiang, Z, Liu, J, Zhang, Y. | | Deposit date: | 2020-05-26 | | Release date: | 2020-07-01 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of inactive and active GABABreceptor.
Cell Res., 30, 2020
|
|
7C7S
 
 | | Cryo-EM structure of the CGP54626-bound human GABA(B) receptor in inactive state. | | Descriptor: | (R)-(cyclohexylmethyl)[(2S)-3-{[(1S)-1-(3,4-dichlorophenyl)ethyl]amino}-2-hydroxypropyl]phosphinic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Mao, C, Shen, C, Li, C, Shen, D, Xu, C, Zhang, S, Zhou, R, Shen, Q, Chen, L, Jiang, Z, Liu, J, Zhang, Y. | | Deposit date: | 2020-05-26 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of inactive and active GABABreceptor.
Cell Res., 30, 2020
|
|
7EB6
 
 | | Crystal structure of GTP-binding protein-like domain of AGAP1 | | Descriptor: | Arf-GAP with GTPase, ANK repeat and PH domain-containing protein 1 | | Authors: | Cheng, N, Zhang, H, Zhang, S, Ma, X, Meng, G. | | Deposit date: | 2021-03-09 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.014 Å) | | Cite: | Crystal structure of the GTP-binding protein-like domain of AGAP1.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7X4B
 
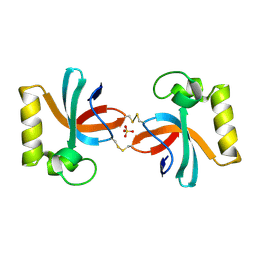 | | Crystal Structure of An Anti-CRISPR Protein | | Descriptor: | Anti-CRISPR protein (AcrIIC1), SULFATE ION | | Authors: | Hu, J, Zhang, S, Gao, J.Y, Liu, X, Liu, J. | | Deposit date: | 2022-03-02 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | A redox switch regulates the assembly and anti-CRISPR activity of AcrIIC1.
Nat Commun, 13, 2022
|
|
7BPI
 
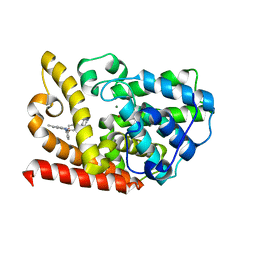 | | The crystal structue of PDE10A complexed with 14 | | Descriptor: | 8-[(E)-2-[5-methyl-1-[3-[3-(4-methylpiperazin-1-yl)propoxy]phenyl]benzimidazol-2-yl]ethenyl]quinoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Yang, Y, Zhang, S, Zhou, Q, Huang, Y.-Y, Guo, L, Luo, H.-B. | | Deposit date: | 2020-03-22 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4000864 Å) | | Cite: | Discovery of highly selective and orally available benzimidazole-based phosphodiesterase 10 inhibitors with improved solubility and pharmacokinetic properties for treatment of pulmonary arterial hypertension.
Acta Pharm Sin B, 10, 2020
|
|
8HJF
 
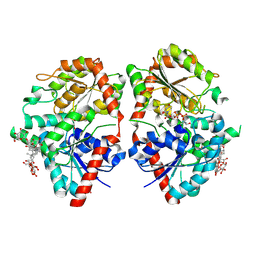 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with M5, state 2 | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, (2S,3S,4S,5R,6R)-2-(hydroxymethyl)-6-[[(2R,3S,4S,5R,6R)-6-[[(3S,8S,9R,10R,11R,13R,14S,17R)-17-[(2S,5R)-5-[(2S,3R,4S,5S,6R)-3-[(2R,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-6-[[(2R,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxymethyl]-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-methyl-6-oxidanyl-heptan-2-yl]-4,4,9,13,14-pentamethyl-11-oxidanyl-2,3,7,8,10,11,12,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-3-yl]oxy]-3,4,5-tris(oxidanyl)oxan-2-yl]methoxy]oxane-3,4,5-triol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
8HJP
 
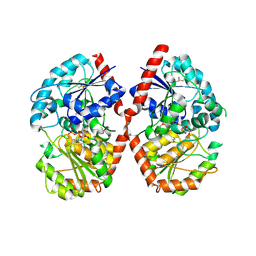 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with UDP state 1 | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, URIDINE-5'-DIPHOSPHATE, glycosyltranseferease | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
8HJN
 
 | |
8HJQ
 
 | |
8HJO
 
 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with UDP state 2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
8HJG
 
 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with M5, state 1 | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, (2S,3S,4S,5R,6R)-2-(hydroxymethyl)-6-[[(2R,3S,4S,5R,6R)-6-[[(3S,8S,9R,10R,11R,13R,14S,17R)-17-[(2S,5R)-5-[(2S,3R,4S,5S,6R)-3-[(2R,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-6-[[(2R,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxymethyl]-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-methyl-6-oxidanyl-heptan-2-yl]-4,4,9,13,14-pentamethyl-11-oxidanyl-2,3,7,8,10,11,12,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-3-yl]oxy]-3,4,5-tris(oxidanyl)oxan-2-yl]methoxy]oxane-3,4,5-triol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
8HJL
 
 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with M3E | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, (2R,3S,4S,5R,6R)-2-(hydroxymethyl)-6-[[(3S,8S,9R,10S,11S,13R,14S,17S)-17-[(2S,5R)-5-[(2S,3R,4S,5R,6S)-6-(hydroxymethyl)-3-[(2S,3R,4S,5S,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-methyl-6-oxidanyl-heptan-2-yl]-4,4,9,13,14-pentamethyl-11-oxidanyl-2,3,7,8,10,11,12,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-3-yl]oxy]oxane-3,4,5-triol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
8HJH
 
 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with SIA, state 3 | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, (2S,3S,4S,5R,6R)-2-(hydroxymethyl)-6-[[(2S,3S,4S,5R,6S)-5-[(2S,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-6-[(3R,6S)-6-[(3S,8S,9R,10R,11S,13R,14S,17R)-3-[(2S,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-4,4,9,13,14-pentamethyl-11-oxidanyl-2,3,7,8,10,11,12,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-17-yl]-2-methyl-2-oxidanyl-heptan-3-yl]oxy-3,4-bis(oxidanyl)oxan-2-yl]methoxy]oxane-3,4,5-triol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
8HJK
 
 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with SIA, state 1 | | Descriptor: | (2S,3S,4S,5R,6R)-2-(hydroxymethyl)-6-[[(2S,3S,4S,5R,6S)-5-[(2S,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-6-[(3R,6S)-6-[(3S,8S,9R,10R,11S,13R,14S,17R)-3-[(2S,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-4,4,9,13,14-pentamethyl-11-oxidanyl-2,3,7,8,10,11,12,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-17-yl]-2-methyl-2-oxidanyl-heptan-3-yl]oxy-3,4-bis(oxidanyl)oxan-2-yl]methoxy]oxane-3,4,5-triol, URIDINE-5'-DIPHOSPHATE, glycosyltranseferease | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.833 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
8J66
 
 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with M3, state 2 | | Descriptor: | (2S,3S,4S,5R,6R)-2-(hydroxymethyl)-6-[[(2R,3S,4S,5R,6S)-6-[(3R,6S)-6-[(3S,8S,9R,10R,11S,13R,14S,17S)-3-[(2R,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-4,4,9,13,14-pentamethyl-11-oxidanyl-2,3,7,8,10,11,12,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-17-yl]-2-methyl-2-oxidanyl-heptan-3-yl]oxy-3,4,5-tris(oxidanyl)oxan-2-yl]methoxy]oxane-3,4,5-triol, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2023-04-24 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
7VUL
 
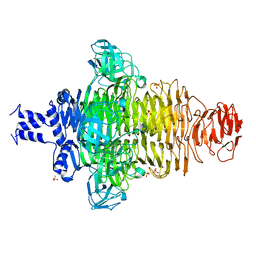 | |
7C2S
 
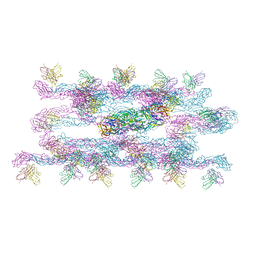 | | Helical reconstruction of Dengue virus serotype 3 complexed with Fab C10 | | Descriptor: | Heavy chain of Fab C10, envelope protein, light chain of Fab C10 | | Authors: | Morrone, S, Chew, S.V, Lim, X.N, Ng, T.S, Kostyuchenko, V.A, Zhang, S, Lok, S.M. | | Deposit date: | 2020-05-09 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (10.4 Å) | | Cite: | High flavivirus structural plasticity demonstrated by a non-spherical morphological variant.
Nat Commun, 11, 2020
|
|
7C2T
 
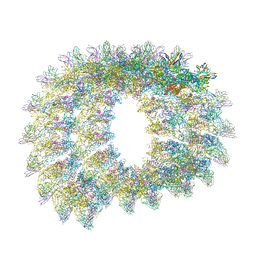 | | Helical reconstruction of Zika virus complexed with Fab C10 | | Descriptor: | Heavy chain from Fab C10, Light chain from Fab C10, M protein, ... | | Authors: | Morrone, S, Chew, S.V, Lim, X.N, Ng, T.S, Kostyuchenko, V.A, Zhang, S, Lok, S.M. | | Deposit date: | 2020-05-09 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | High flavivirus structural plasticity demonstrated by a non-spherical morphological variant.
Nat Commun, 11, 2020
|
|
7KZ7
 
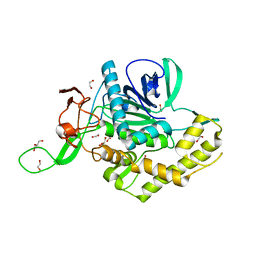 | | Crystals Structure of the Mutated Protease Domain of Botulinum Neurotoxin X (X4130B1). | | Descriptor: | 1,2-ETHANEDIOL, Botulinum neurotoxin type X, GLYCEROL, ... | | Authors: | Blum, T.R, Liu, H, Packer, M.S, Xiong, X, Lee, P.G, Zhang, S, Richter, M, Minasov, G, Satchell, K.J.F, Dong, M, Liu, D.R, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Phage-assisted evolution of botulinum neurotoxin proteases with reprogrammed specificity.
Science, 371, 2021
|
|
7CKR
 
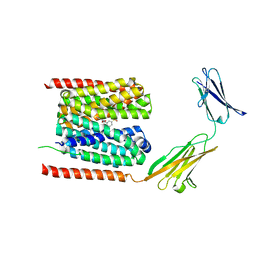 | | Cryo-EM structure of the human MCT1/Basigin-2 complex in the presence of anti-cancer drug candidate BAY-8002 in the outward-open conformation. | | Descriptor: | 2-[[2-chloranyl-5-(phenylsulfonyl)phenyl]carbonylamino]benzoic acid, Basigin, Monocarboxylate transporter 1 | | Authors: | Wang, N, Jiang, X, Zhang, S, Zhu, A, Yuan, Y, Lei, J, Yan, C. | | Deposit date: | 2020-07-18 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates.
Cell, 184, 2021
|
|
7CKO
 
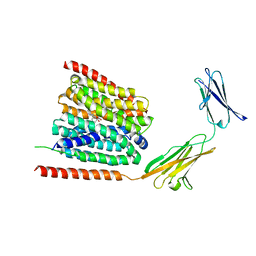 | | Cryo-EM structure of the human MCT1/Basigin-2 complex in the presence of anti-cancer drug candidate 7ACC2 in the inward-open conformation | | Descriptor: | 7-[methyl-(phenylmethyl)amino]-2-oxidanylidene-chromene-3-carboxylic acid, Basigin, Monocarboxylate transporter 1 | | Authors: | Wang, N, Jiang, X, Zhang, S, Zhu, A, Yuan, Y, Lei, J, Yan, C. | | Deposit date: | 2020-07-18 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates.
Cell, 184, 2021
|
|
7DA5
 
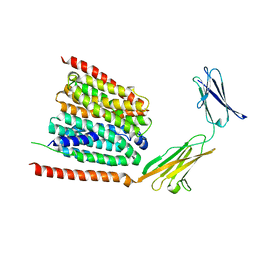 | | Cryo-EM structure of the human MCT1 D309N mutant in complex with Basigin-2 in the inward-open conformation. | | Descriptor: | Basigin, Monocarboxylate transporter 1 | | Authors: | Wang, N, Jiang, X, Zhang, S, Zhu, A, Yuan, Y, Lei, J, Yan, C. | | Deposit date: | 2020-10-14 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates.
Cell, 184, 2021
|
|
7V5K
 
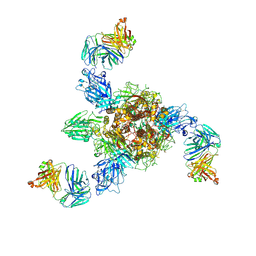 | | MERS S ectodomain trimer in complex with neutralizing antibody 0722 (state 1) | | Descriptor: | 0722 H, 0722 L, Spike glycoprotein | | Authors: | Wang, X, Zhao, J, Wang, Z, Zeng, J, Zhang, S, Wang, Y. | | Deposit date: | 2021-08-17 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | MERS S ectodomain trimer in complex with neutralizing antibody 0722 (state 1)
to be published
|
|
7V5J
 
 | | MERS S ectodomain trimer in complex with neutralizing antibody 0722(state 2) | | Descriptor: | 0722 H, 0722 L, Spike glycoprotein | | Authors: | Wang, X, Zhao, J, Wang, Z, Zeng, J, Zhang, S, Wang, Y. | | Deposit date: | 2021-08-17 | | Release date: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | MERS S ectodomain trimer in complex with neutralizing antibody 0722(state 2)
to be published
|
|
