7U13
 
 | | TMEM106B(120-254) singlet amyloid fibril from frontotemporal lobar degeneration with TDP-43 pathology (FTLD-TDP) type A (case 4) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Fitzpatrick, A.W.P, Stowell, M.H.B, Chang, A, Xiang, X, Wang, J, Lee, C, Arakhamia, T, Simjanoska, M, Wang, C, Carlomagno, Y, Zhang, G, Dhingra, S, Thierry, M, Perneel, J, Heeman, B, Forgrave, L.M, DeTure, M, DeMarco, M.L, Cook, C.N, Rademakers, R, Dickson, D, Petrucelli, L, Mackenzie, I.R.A. | | Deposit date: | 2022-02-19 | | Release date: | 2022-03-23 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Homotypic fibrillization of TMEM106B across diverse neurodegenerative diseases.
Cell, 185, 2022
|
|
7EVJ
 
 | | Crystal structure of CBP bromodomain liganded with 9c | | Descriptor: | 3-acetyl-1-((3-(1-cyclopropyl-1H-pyrazol-4-yl)-2-fluoro-5-(hydroxymethyl)phenyl)carbamoyl)indolizin-7-yl dimethylcarbamate, CREB-binding protein, GLYCEROL, ... | | Authors: | Xiang, Q, Wang, C, Wu, T, Zhang, Y, Zhang, C, Luo, G, Wu, X, Shen, H, Xu, Y. | | Deposit date: | 2021-05-21 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 1-(Indolizin-3-yl)ethan-1-ones as CBP Bromodomain Inhibitors for the Treatment of Prostate Cancer.
J.Med.Chem., 65, 2022
|
|
4X9Z
 
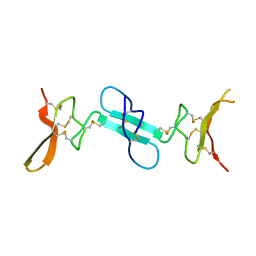 | | Dimeric conotoxin alphaD-GeXXA | | Descriptor: | alphaD-conotoxin GeXXA from the venom of Conus generalis | | Authors: | Xu, S, Zhang, T, Kompella, S, Adams, D, Ding, J, Wang, C. | | Deposit date: | 2014-12-12 | | Release date: | 2015-12-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conotoxin alpha D-GeXXA utilizes a novel strategy to antagonize nicotinic acetylcholine receptors
Sci Rep, 5, 2015
|
|
3WUY
 
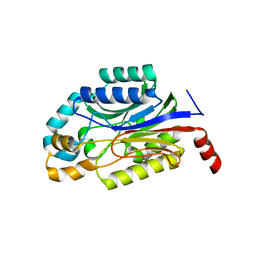 | | Crystal structure of Nit6803 | | Descriptor: | Nitrilase | | Authors: | Yuan, Y.A, Yin, B, Wang, C. | | Deposit date: | 2014-05-09 | | Release date: | 2014-12-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural insights into enzymatic activity and substrate specificity determination by a single amino acid in nitrilase from Syechocystis sp. PCC6803
J.Struct.Biol., 188, 2014
|
|
4ZA1
 
 | | Crystal Structure of NosA Involved in Nosiheptide Biosynthesis | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, NosA | | Authors: | Liu, S, Guo, H, Zhang, T, Han, L, Yao, P, Zhang, Y, Rong, N, Yu, Y, Lan, W, Wang, C, Ding, J, Wang, R, Liu, W, Cao, C. | | Deposit date: | 2015-04-13 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based Mechanistic Insights into Terminal Amide Synthase in Nosiheptide-Represented Thiopeptides Biosynthesis
Sci Rep, 5, 2015
|
|
4NC3
 
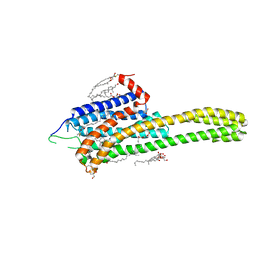 | | Crystal structure of the 5-HT2B receptor solved using serial femtosecond crystallography in lipidic cubic phase. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHOLESTEROL, ... | | Authors: | Liu, W, Wacker, D, Gati, C, Han, G.W, James, D, Wang, D, Nelson, G, Weierstall, U, Katritch, V, Barty, A, Zatsepin, N.A, Li, D, Messerschmidt, M, Boutet, S, Williams, G.J, Koglin, J.E, Seibert, M.M, Wang, C, Shah, S.T.A, Basu, S, Fromme, R, Kupitz, C, Rendek, K.N, Grotjohann, I, Fromme, P, Kirian, R.A, Beyerlein, K.R, White, T.A, Chapman, H.N, Caffrey, M, Spence, J.C.H, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Serial femtosecond crystallography of G protein-coupled receptors.
Science, 342, 2013
|
|
4XHR
 
 | | Structure of a phospholipid trafficking complex, native | | Descriptor: | Mitochondrial distribution and morphology protein 35, Protein UPS1, mitochondrial | | Authors: | Yu, F, He, F, Wang, C, Zhang, P. | | Deposit date: | 2015-01-06 | | Release date: | 2015-07-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of intramitochondrial phosphatidic acid transport mediated by Ups1-Mdm35 complex
Embo Rep., 16, 2015
|
|
4XIZ
 
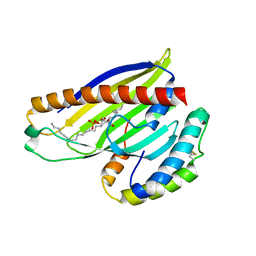 | | Structure of a phospholipid trafficking complex with substrate | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, Mitochondrial distribution and morphology protein 35, Protein UPS1, ... | | Authors: | Yu, F, He, F, Wang, C, Zhang, P. | | Deposit date: | 2015-01-08 | | Release date: | 2015-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of intramitochondrial phosphatidic acid transport mediated by Ups1-Mdm35 complex
Embo Rep., 16, 2015
|
|
2WPV
 
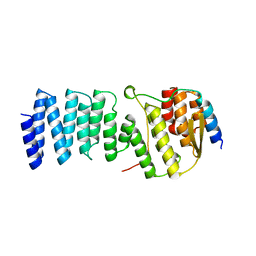 | | Crystal structure of S. cerevisiae Get4-Get5 complex | | Descriptor: | MERCURY (II) ION, UBIQUITIN-LIKE PROTEIN MDY2, UPF0363 PROTEIN YOR164C | | Authors: | Chang, Y.-W, Chuang, Y.-C, Ho, Y.-C, Cheng, M.-Y, Sun, Y.-J, Hsiao, C.-D, Wang, C. | | Deposit date: | 2009-08-11 | | Release date: | 2010-01-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure of Get4-Get5 Complex and its Interactions with Sgt2, Get3, and Ydj1.
J.Biol.Chem., 285, 2010
|
|
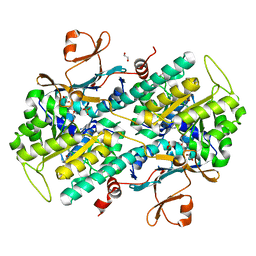
4N9B
 
 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1-methyl-N-(pyridin-3-yl)-1H-pyrazole-5-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhai, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | Deposit date: | 2013-10-20 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.859 Å) | | Cite: | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4N9D
 
 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1,2-ETHANEDIOL, 4-({[(4-tert-butylphenyl)sulfonyl]amino}methyl)-N-(pyridin-3-yl)benzamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhao, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | Deposit date: | 2013-10-20 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4N9C
 
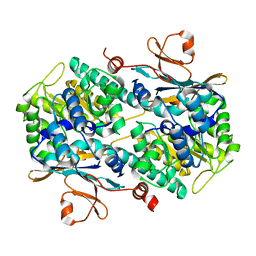 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 5-nitro-1H-benzimidazole, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhao, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | Deposit date: | 2013-10-20 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4N9E
 
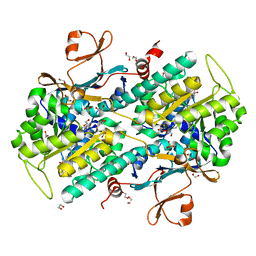 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1-benzoylpiperidin-4-yl)methyl]-N-(pyridin-3-yl)-1H-benzimidazole-5-carboxamide, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ... | | Authors: | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhao, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | Deposit date: | 2013-10-20 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
7EP7
 
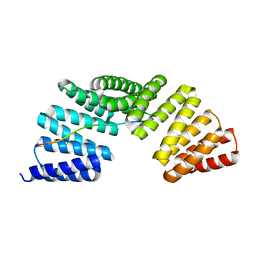 | | The complex structure of Gpsm2 and Whirlin | | Descriptor: | G-protein-signaling modulator 2, Whirlin | | Authors: | Lin, L, Shi, Y, Wang, C, Zhu, J. | | Deposit date: | 2021-04-26 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Promotion of row 1-specific tip complex condensates by Gpsm2-G alpha i provides insights into row identity of the tallest stereocilia.
Sci Adv, 8, 2022
|
|
6KZJ
 
 | | Crystal structure of Ankyrin B/NdeL1 complex | | Descriptor: | Ankyrin-2, Nuclear distribution protein nudE-like 1 | | Authors: | Ye, J, Li, J, Ye, F, Zhang, M, Zhang, Y, Wang, C. | | Deposit date: | 2019-09-24 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic insights into the interactions of dynein regulator Ndel1 with neuronal ankyrins and implications in polarity maintenance.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7ERL
 
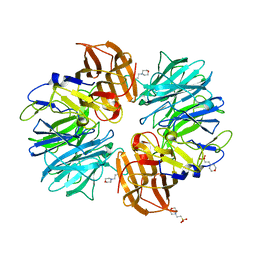 | |
5YQX
 
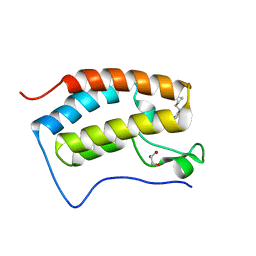 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | (2R)-2-(cyclopropylmethyl)-7-(3,5-dimethyl-1,2-oxazol-4-yl)-4H-1,4-benzoxazin-3-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Xue, X, Zhang, Y, Wang, C, Song, M. | | Deposit date: | 2017-11-08 | | Release date: | 2018-11-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Benzoxazinone-containing 3,5-dimethylisoxazole derivatives as BET bromodomain inhibitors for treatment of castration-resistant prostate cancer.
Eur.J.Med.Chem., 152, 2018
|
|
6LOD
 
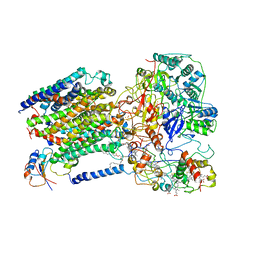 | | Cryo-EM structure of the air-oxidized photosynthetic alternative complex III from Roseiflexus castenholzii | | Descriptor: | Cytochrome c domain-containing protein, FE3-S4 CLUSTER, Fe-S-cluster-containing hydrogenase components 1-like protein, ... | | Authors: | Shi, Y, Xin, Y.Y, Wang, C, Blankenship, R.E, Sun, F, Xu, X.L. | | Deposit date: | 2020-01-05 | | Release date: | 2020-11-04 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of the air-oxidized and dithionite-reduced photosynthetic alternative complex III from Roseiflexus castenholzii .
Sci Adv, 6, 2020
|
|
6LOE
 
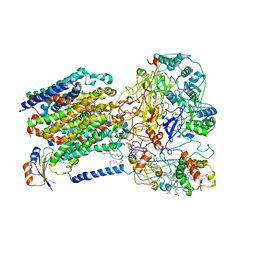 | | Cryo-EM structure of the dithionite-reduced photosynthetic alternative complex III from Roseiflexus castenholzii | | Descriptor: | Cytochrome c domain-containing protein, FE3-S4 CLUSTER, Fe-S-cluster-containing hydrogenase components 1-like protein, ... | | Authors: | Shi, Y, Xin, Y.Y, Wang, C, Blankenship, R.E, Sun, F, Xu, X.L. | | Deposit date: | 2020-01-05 | | Release date: | 2020-11-04 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of the air-oxidized and dithionite-reduced photosynthetic alternative complex III from Roseiflexus castenholzii .
Sci Adv, 6, 2020
|
|
9JM0
 
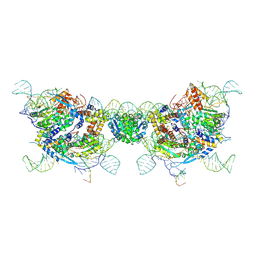 | | retron Ec86-effector fiber | | Descriptor: | DNA (85-MER), NICOTINAMIDE, RNA (5'-R(P*CP*GP*UP*AP*AP*GP*GP*GP*UP*GP*CP*GP*CP*A)-3'), ... | | Authors: | Wang, Y.J, Guan, Z.Y, Wang, C, Zou, T.T. | | Deposit date: | 2024-09-20 | | Release date: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | DNA methylation activates retron Ec86 filaments for antiphage defense.
Cell Rep, 43, 2024
|
|
4O1I
 
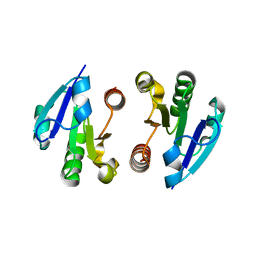 | | Crystal Structure of the regulatory domain of MtbGlnR | | Descriptor: | Transcriptional regulatory protein | | Authors: | Lin, W, Wang, C, Zhang, P. | | Deposit date: | 2013-12-16 | | Release date: | 2014-04-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atypical OmpR/PhoB Subfamily Response Regulator GlnR of Actinomycetes Functions as a Homodimer, Stabilized by the Unphosphorylated Conserved Asp-focused Charge Interactions
J.Biol.Chem., 289, 2014
|
|
4O1H
 
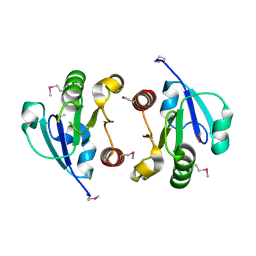 | | Crystal Structure of the regulatory domain of AmeGlnR | | Descriptor: | Transcription regulator GlnR | | Authors: | Lin, W, Wang, C, Zhang, P. | | Deposit date: | 2013-12-16 | | Release date: | 2014-04-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atypical OmpR/PhoB Subfamily Response Regulator GlnR of Actinomycetes Functions as a Homodimer, Stabilized by the Unphosphorylated Conserved Asp-focused Charge Interactions
J.Biol.Chem., 289, 2014
|
|
6NZS
 
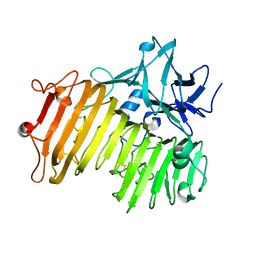 | | Dextranase AoDex KQ11 | | Descriptor: | Dextranase | | Authors: | Ren, W, Yan, W, Gu, L, Feng, Y, Dong, D, Wang, S, Wang, C, Lyu, M. | | Deposit date: | 2019-02-14 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus
To Be Published
|
|
5Y8Y
 
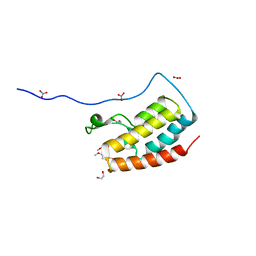 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromanyl-2-methoxy-N-(6-methoxy-3-methyl-1,2-benzoxazol-5-yl)benzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-21 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
5EQJ
 
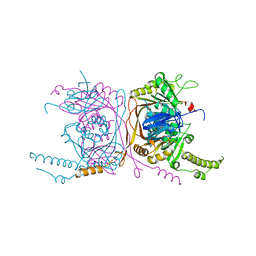 | | Crystal structure of the two-subunit tRNA m1A58 methyltransferase from Saccharomyces cerevisiae | | Descriptor: | tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRM61, tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit TRM6 | | Authors: | Zhu, Y, Wang, M, Wang, C, Fan, X, Jiang, X, Teng, M, Li, X. | | Deposit date: | 2015-11-13 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the two-subunit tRNA m(1)A58 methyltransferase TRM6-TRM61 from Saccharomyces cerevisiae.
Sci Rep, 6, 2016
|
|
