6H4V
 
 | | Crystal structure of human KDM4A in complex with compound 34g | | Descriptor: | 8-[4-(1-cyclopentylpiperidin-4-yl)pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | C8-substituted pyrido[3,4-d]pyrimidin-4(3H)-ones: Studies towards the identification of potent, cell penetrant Jumonji C domain containing histone lysine demethylase 4 subfamily (KDM4) inhibitors, compound profiling in cell-based target engagement assays.
Eur.J.Med.Chem., 177, 2019
|
|
6H4T
 
 | | Crystal structure of human KDM4A in complex with compound 19a | | Descriptor: | 8-[4-(2-spiro[1,2-dihydroindene-3,4'-piperidine]-1'-ylethyl)pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | C8-substituted pyrido[3,4-d]pyrimidin-4(3H)-ones: Studies towards the identification of potent, cell penetrant Jumonji C domain containing histone lysine demethylase 4 subfamily (KDM4) inhibitors, compound profiling in cell-based target engagement assays.
Eur.J.Med.Chem., 177, 2019
|
|
6H4Q
 
 | | Crystal structure of human KDM4A in complex with compound 34a | | Descriptor: | 8-[4-(1-methylpiperidin-4-yl)pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | C8-substituted pyrido[3,4-d]pyrimidin-4(3H)-ones: Studies towards the identification of potent, cell penetrant Jumonji C domain containing histone lysine demethylase 4 subfamily (KDM4) inhibitors, compound profiling in cell-based target engagement assays.
Eur.J.Med.Chem., 177, 2019
|
|
6H4W
 
 | | Crystal structure of human KDM4A in complex with compound 19d | | Descriptor: | 8-[4-[2-[4-(3-chlorophenyl)-4-methyl-piperidin-1-yl]ethyl]pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | C8-substituted pyrido[3,4-d]pyrimidin-4(3H)-ones: Studies towards the identification of potent, cell penetrant Jumonji C domain containing histone lysine demethylase 4 subfamily (KDM4) inhibitors, compound profiling in cell-based target engagement assays.
Eur.J.Med.Chem., 177, 2019
|
|
6H4X
 
 | | Crystal structure of human KDM4A in complex with compound 17b | | Descriptor: | 8-[4-[2-[4-(4-pyridin-3-ylphenyl)piperidin-1-yl]ethyl]pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | C8-substituted pyrido[3,4-d]pyrimidin-4(3H)-ones: Studies towards the identification of potent, cell penetrant Jumonji C domain containing histone lysine demethylase 4 subfamily (KDM4) inhibitors, compound profiling in cell-based target engagement assays.
Eur.J.Med.Chem., 177, 2019
|
|
6H4U
 
 | | Crystal structure of human KDM4A in complex with compound 34b | | Descriptor: | 8-[4-(1-methylpiperidin-4-yl)pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | C8-substituted pyrido[3,4-d]pyrimidin-4(3H)-ones: Studies towards the identification of potent, cell penetrant Jumonji C domain containing histone lysine demethylase 4 subfamily (KDM4) inhibitors, compound profiling in cell-based target engagement assays.
Eur.J.Med.Chem., 177, 2019
|
|
6H4S
 
 | | Crystal structure of human KDM4A in complex with compound 16m | | Descriptor: | 8-[4-[2-[4-[3-[2-(dimethylamino)ethyl]phenyl]piperidin-1-yl]ethyl]pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | C8-substituted pyrido[3,4-d]pyrimidin-4(3H)-ones: Studies towards the identification of potent, cell penetrant Jumonji C domain containing histone lysine demethylase 4 subfamily (KDM4) inhibitors, compound profiling in cell-based target engagement assays.
Eur.J.Med.Chem., 177, 2019
|
|
4J95
 
 | |
4J97
 
 | |
3F1R
 
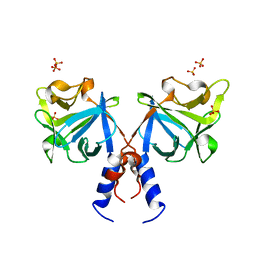 | | Crystal structure of FGF20 dimer | | Descriptor: | Fibroblast growth factor 20, SULFATE ION | | Authors: | Kalinina, J, Mohammadi, M. | | Deposit date: | 2008-10-28 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Homodimerization controls the fibroblast growth factor 9 subfamily's receptor binding and heparan sulfate-dependent diffusion in the extracellular matrix
Mol.Cell.Biol., 29, 2009
|
|
4QQ5
 
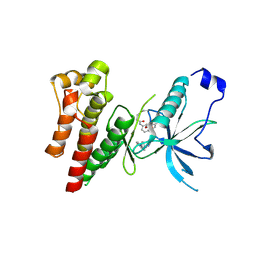 | | Crystal Structure of FGF Receptor (FGFR) 4 Kinase Harboring the V550L Gate-Keeper Mutation in Complex With FIIN-2, an Irreversible Tyrosine Kinase Inhibitor Capable of Overcoming FGFR Kinase Gate-Keeper Mutations | | Descriptor: | Fibroblast growth factor receptor 4, N-(4-{[3-(3,5-dimethoxyphenyl)-7-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-2-oxo-3,4-dihydropyrimido[4,5-d]pyrimidin-1(2H)-yl]methyl}phenyl)propanamide | | Authors: | Huang, Z, Mohammadi, M. | | Deposit date: | 2014-06-26 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | DFG-out Mode of Inhibition by an Irreversible Type-1 Inhibitor Capable of Overcoming Gate-Keeper Mutations in FGF Receptors.
Acs Chem.Biol., 10, 2015
|
|
4QQJ
 
 | |
4QQT
 
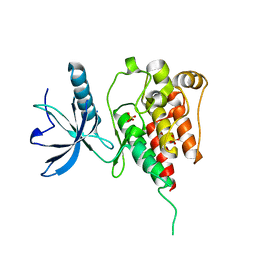 | |
4QRC
 
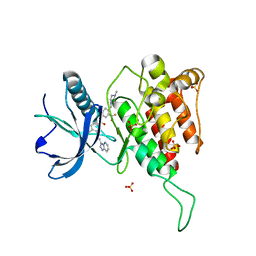 | | Crystal Structure of the Tyrosine Kinase Domain of FGF Receptor 4 in Complex with Ponatinib | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, Fibroblast growth factor receptor 4, SULFATE ION | | Authors: | Huang, Z, Mohammadi, M. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | DFG-out Mode of Inhibition by an Irreversible Type-1 Inhibitor Capable of Overcoming Gate-Keeper Mutations in FGF Receptors.
Acs Chem.Biol., 10, 2015
|
|
4G7E
 
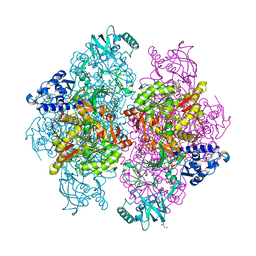 | |
5DFK
 
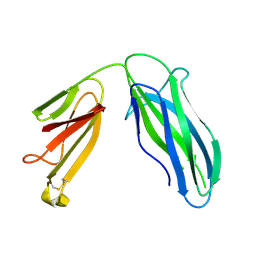 | |
5EG3
 
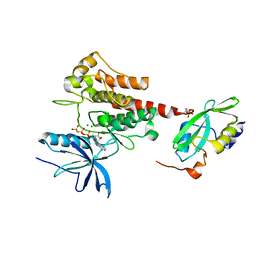 | | Crystal Structure of the Activated FGF Receptor 2 (FGFR2) Kinase Domain in complex with the cSH2 domain of Phospholipase C gamma (PLCgamma) | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1, Fibroblast growth factor receptor 2, MAGNESIUM ION, ... | | Authors: | Huang, Z, Li, X, Mohammadi, M. | | Deposit date: | 2015-10-26 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Two FGF Receptor Kinase Molecules Act in Concert to Recruit and Transphosphorylate Phospholipase C gamma.
Mol.Cell, 61, 2016
|
|
3TTY
 
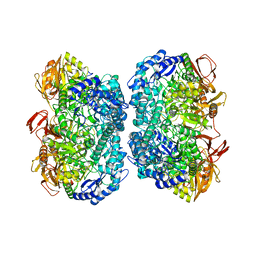 | | Crystal structure of beta-galactosidase from Bacillus circulans sp. alkalophilus in complex with galactose | | Descriptor: | Beta-galactosidase, ZINC ION, alpha-D-galactopyranose | | Authors: | Maksimainen, M, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2011-09-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural analysis, enzymatic characterization, and catalytic mechanisms of beta-galactosidase from Bacillus circulans sp. alkalophilus.
Febs J., 279, 2012
|
|
3TTS
 
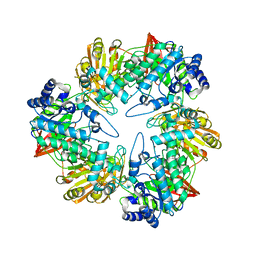 | | Crystal structure of beta-galactosidase from Bacillus circulans sp. alkalophilus | | Descriptor: | Beta-galactosidase, ZINC ION | | Authors: | Maksimainen, M, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2011-09-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural analysis, enzymatic characterization, and catalytic mechanisms of beta-galactosidase from Bacillus circulans sp. alkalophilus.
Febs J., 279, 2012
|
|
5IN3
 
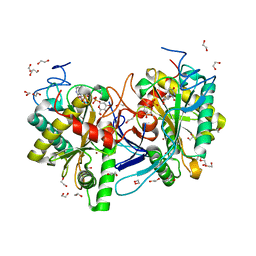 | | Crystal structure of glucose-1-phosphate bound nucleotidylated human galactose-1-phosphate uridylyltransferase | | Descriptor: | 1,2-ETHANEDIOL, 1-O-phosphono-alpha-D-glucopyranose, 5,6-DIHYDROURIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Kopec, J, McCorvie, T, Tallant, C, Velupillai, S, Shrestha, L, Fitzpatrick, F, Patel, D, Chalk, R, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2016-03-07 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Molecular basis of classic galactosemia from the structure of human galactose 1-phosphate uridylyltransferase.
Hum.Mol.Genet., 25, 2016
|
|
1S26
 
 | | Structure of Anthrax Edema Factor-Calmodulin-alpha,beta-methyleneadenosine 5'-triphosphate Complex Reveals an Alternative Mode of ATP Binding to the Catalytic Site | | Descriptor: | CALCIUM ION, Calmodulin, Calmodulin-sensitive adenylate cyclase, ... | | Authors: | Shen, Y, Zhukovskaya, N.L, Bohm, A, Tang, W.-J. | | Deposit date: | 2004-01-08 | | Release date: | 2004-04-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of anthrax edema factor-calmodulin-adenosine-5'-(alpha,beta-methylene)-triphosphate complex reveals an alternative mode of ATP binding to the catalytic site
Biochem.Biophys.Res.Commun., 317, 2004
|
|
5A4R
 
 | | Crystal structure of a vitamin B12 trafficking protein | | Descriptor: | METHYLMALONIC ACIDURIA AND HOMOCYSTINURIA TYPE D HOMOLOG, MITOCHONDRIAL | | Authors: | Kopec, J, Fitzpatrick, F, Froese, D.S, Velupillai, S, Nowak, R, Chalk, R, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Fowler, B, Baumgartner, M.R, Yue, W.W. | | Deposit date: | 2015-06-11 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Insights Into the Mmachc-Mmadhc Protein Complex Involved in Vitamin B12 Trafficking.
J.Biol.Chem., 290, 2015
|
|
