3WGL
 
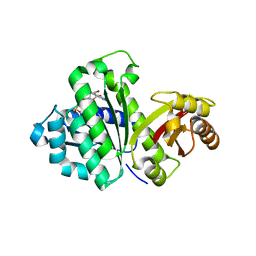 | | STAPHYLOCOCCUS AUREUS FTSZ T7 mutant substituted for GAN bound with GDP, DeltaT7GAN-GDP | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Han, X, Matsui, T, Yu, J, Tanaka, I, Yao, M. | | Deposit date: | 2013-08-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.066 Å) | | Cite: | Structural change in FtsZ Induced by intermolecular interactions between bound GTP and the T7 loop
J.Biol.Chem., 289, 2014
|
|
7F3P
 
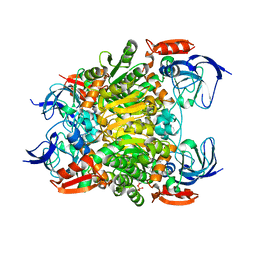 | | Crystal structure of a nadp-dependent alcohol dehydrogenase mutant in apo form | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent isopropanol dehydrogenase, ZINC ION | | Authors: | Han, X, Bi, Y, Wei, H.L, Gao, J, Li, Q, Qu, G, Sun, Z.T, Liu, W.D. | | Deposit date: | 2021-06-16 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Unlocking the Stereoselectivity and Substrate Acceptance of Enzymes: Proline-Induced Loop Engineering Test.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7OZR
 
 | |
6H6D
 
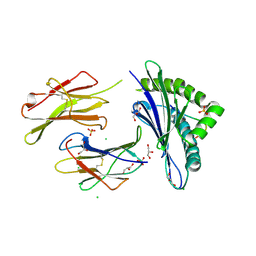 | |
7W1I
 
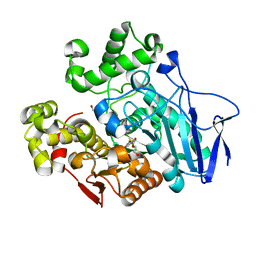 | | Crystal structure of carboxylesterase mutant from Thermobifida fusca with C8X and C9C | | Descriptor: | 4-(2-hydroxyethyloxycarbonyl)benzoic acid, Carboxylesterase, bis(2-hydroxyethyl) benzene-1,4-dicarboxylate | | Authors: | Han, X, Gerlis, H, Li, Z, Gao, J, Wei, R, Liu, W. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural Insights into (Tere)phthalate-Ester Hydrolysis by a Carboxylesterase and Its Role in Promoting PET Depolymerization
Acs Catalysis, 12, 2022
|
|
7W1K
 
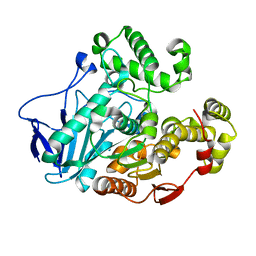 | | Crystal structure of carboxylesterase from Thermobifida fusca | | Descriptor: | Carboxylesterase | | Authors: | Han, X, Gerlis, H, Li, Z, Gao, J, Wei, R, Liu, W. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural Insights into (Tere)phthalate-Ester Hydrolysis by a Carboxylesterase and Its Role in Promoting PET Depolymerization
Acs Catalysis, 12, 2022
|
|
7W1J
 
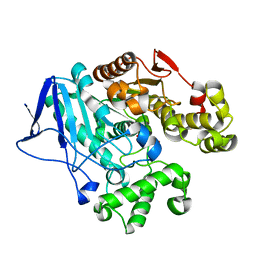 | | Crystal structure of carboxylesterase from Thermobifida fusca with J1K | | Descriptor: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, Carboxylesterase | | Authors: | Han, X, Gerlis, H, Li, Z, Gao, J, Wei, R, Liu, W. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Insights into (Tere)phthalate-Ester Hydrolysis by a Carboxylesterase and Its Role in Promoting PET Depolymerization
Acs Catalysis, 12, 2022
|
|
7W1L
 
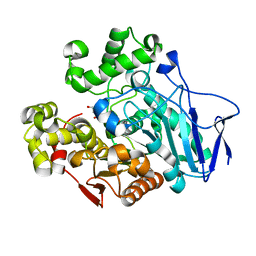 | | Crystal structure of carboxylesterase mutant from Thermobifida fusca with C8X | | Descriptor: | Carboxylesterase, bis(2-hydroxyethyl) benzene-1,4-dicarboxylate | | Authors: | Han, X, Gerlis, H, Li, Z, Gao, J, Wei, R, Liu, W. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural Insights into (Tere)phthalate-Ester Hydrolysis by a Carboxylesterase and Its Role in Promoting PET Depolymerization
Acs Catalysis, 12, 2022
|
|
6X19
 
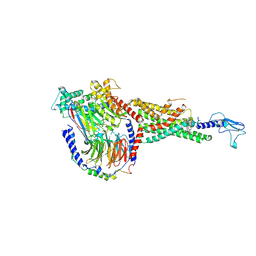 | | Non peptide agonist CHU-128, bound to Glucagon-Like peptide-1 (GLP-1) Receptor | | Descriptor: | 3-{(1S)-1-[5-(2,2-dimethylmorpholin-4-yl)-2-{(4S)-2-(4-fluoro-3,5-dimethylphenyl)-3-[3-(4-fluoro-1-methyl-1H-indazol-5-yl)-2-oxo-2,3-dihydro-1H-imidazol-1-yl]-4-methyl-2,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carbonyl}-1H-indol-1-yl]butyl}-1,2,4-oxadiazol-5(4H)-one, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Belousoff, M.J, Zhang, X, Danev, R. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-09 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Differential GLP-1R Binding and Activation by Peptide and Non-peptide Agonists.
Mol.Cell, 80, 2020
|
|
7P51
 
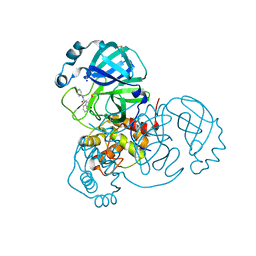 | | CRYSTAL STRUCTURE OF THE SARS-COV-2 MAIN PROTEASE COMPLEXED WITH FRAGMENT F01 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(5-chloropyridin-2-yl)-3-oxo-2,3-dihydro-1H-indene-1-carboxamide, ... | | Authors: | Hanoulle, X, Moschidi, D. | | Deposit date: | 2021-07-13 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.474 Å) | | Cite: | NMR Spectroscopy of the Main Protease of SARS-CoV-2 and Fragment-Based Screening Identify Three Protein Hotspots and an Antiviral Fragment.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6X18
 
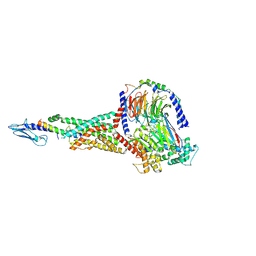 | | GLP-1 peptide hormone bound to Glucagon-Like peptide-1 (GLP-1) Receptor | | Descriptor: | Glucagon, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Belousoff, M.J, Zhang, X, Danev, R. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-09 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Differential GLP-1R Binding and Activation by Peptide and Non-peptide Agonists.
Mol.Cell, 80, 2020
|
|
6X1A
 
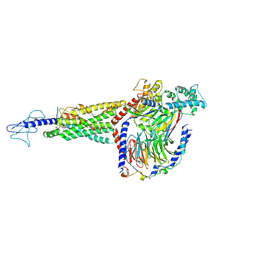 | | Non peptide agonist PF-06882961, bound to Glucagon-Like peptide-1 (GLP-1) Receptor | | Descriptor: | 2-[(4-{6-[(4-cyano-2-fluorophenyl)methoxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-{[(2S)-oxetan-2-yl]methyl}-1H-benzimidazole-6-carboxylic acid, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Belousoff, M.J, Zhang, X, Danev, R. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-09 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Differential GLP-1R Binding and Activation by Peptide and Non-peptide Agonists.
Mol.Cell, 80, 2020
|
|
3O5X
 
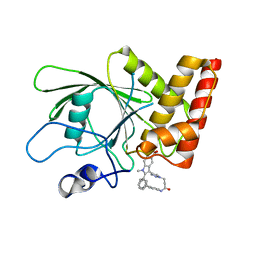 | | Crystal structure of the oncogenic tyrosine phosphatase SHP2 complexed with a salicylic acid-based small molecule inhibitor | | Descriptor: | 3-{1-[3-(biphenyl-4-ylamino)-3-oxopropyl]-1H-1,2,3-triazol-4-yl}-6-hydroxy-1-methyl-2-phenyl-1H-indole-5-carboxylic acid, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Zhang, Z.-Y, Zhang, X, He, Y, Liu, S, Yu, Z, Jiang, Z, Yang, Z, Dong, Y, Nabinger, S.C, Wu, L, Gunawan, A.M, Wang, L, Chan, R.J. | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Salicylic acid based small molecule inhibitor for the oncogenic Src homology-2 domain containing protein tyrosine phosphatase-2 (SHP2).
J.Med.Chem., 53, 2010
|
|
8HK1
 
 | | The cryo-EM structure of human pre-17S U2 snRNP | | Descriptor: | ATP-dependent RNA helicase DDX42, HIV Tat-specific factor 1, PHD finger-like domain-containing protein 5A, ... | | Authors: | Zhang, X, Zhan, X, Shi, Y. | | Deposit date: | 2022-11-24 | | Release date: | 2023-03-08 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanisms of the RNA helicases DDX42 and DDX46 in human U2 snRNP assembly.
Nat Commun, 14, 2023
|
|
7YSK
 
 | | Crystal structure of D-Cysteine desulfhydrase from Pectobacterium atrosepticum | | Descriptor: | D-Cysteine desulfhydrase | | Authors: | Zhang, X, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-08-12 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization and structural basis of D-cysteine desulfhydrase from Pectobacterium atrosepticum
Tetrahedron, 2022
|
|
7YSL
 
 | | Crystal structure of D-Cysteine desulfhydrase with a trapped PLP-pyruvate geminal diamine | | Descriptor: | 1,2-ETHANEDIOL, D-Cysteine desulfhydrase, FORMIC ACID | | Authors: | Zhang, X, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-08-12 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Characterization and structural basis of D-cysteine desulfhydrase from Pectobacterium atrosepticum
Tetrahedron, 2022
|
|
8WCN
 
 | | Cryo-EM structure of PAO1-ImcA with GMPCPP | | Descriptor: | Diguanylate cyclase, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Zhan, X.L, Zhang, K, Wang, C.C, Fan, Q, Tang, X.J, Zhang, X, Wang, K, Fu, Y, Liang, H.H. | | Deposit date: | 2023-09-13 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A c-di-GMP signaling module controls responses to iron in Pseudomonas aeruginosa.
Nat Commun, 15, 2024
|
|
5FB1
 
 | | Crystal Structure of a PHD finger bound to histone H3 K9me3 peptide | | Descriptor: | MALONATE ION, Nuclear autoantigen Sp-100, Peptide from Histone H3, ... | | Authors: | Li, H, Zhang, X. | | Deposit date: | 2015-12-13 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Multifaceted Histone H3 Methylation and Phosphorylation Readout by the Plant Homeodomain Finger of Human Nuclear Antigen Sp100C
J.Biol.Chem., 291, 2016
|
|
5FB0
 
 | | Crystal Structure of a PHD finger bound to histone H3 T3ph peptide | | Descriptor: | Nuclear autoantigen Sp-100, Peptide from Histone H3, ZINC ION | | Authors: | Li, H, Zhang, X. | | Deposit date: | 2015-12-13 | | Release date: | 2016-05-04 | | Last modified: | 2016-06-22 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Multifaceted Histone H3 Methylation and Phosphorylation Readout by the Plant Homeodomain Finger of Human Nuclear Antigen Sp100C
J.Biol.Chem., 291, 2016
|
|
5GQH
 
 | | Cryo-EM structure of PaeCas3-AcrF3 complex | | Descriptor: | CRISPR-associated nuclease/helicase Cas3 subtype I-F/YPEST, anti-CRISPR protein 3 | | Authors: | Zhang, X, Ma, J, Wang, Y, Wang, J. | | Deposit date: | 2016-08-07 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A CRISPR evolutionary arms race: structural insights into viral anti-CRISPR/Cas responses
Cell Res., 26, 2016
|
|
7WOS
 
 | | The state 3 of Omicron Spike with bispecific antibody FD01 | | Descriptor: | 16L9 Fv, 2-acetamido-2-deoxy-beta-D-glucopyranose, GW01 Fv, ... | | Authors: | Zhang, X, Zhan, W.Q, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Combating the SARS-CoV-2 Omicron (BA.1) and BA.2 with potent bispecific antibodies engineered from non-Omicron neutralizing antibodies
Cell Discov, 8, 2022
|
|
7WOQ
 
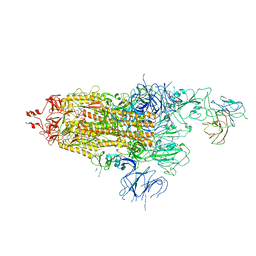 | | The state 1 of Omicron Spike with bispecific antibody FD01 | | Descriptor: | 16L9 Fv, 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Zhan, W.Q, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Combating the SARS-CoV-2 Omicron (BA.1) and BA.2 with potent bispecific antibodies engineered from non-Omicron neutralizing antibodies
Cell Discov, 8, 2022
|
|
7WOU
 
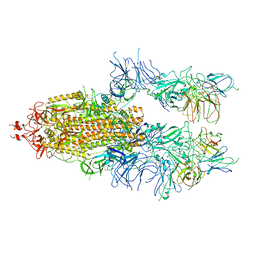 | | The state 4 of Omicron Spike with bispecific antibody FD01 | | Descriptor: | 16L9 Fv, 2-acetamido-2-deoxy-beta-D-glucopyranose, GW01 Fv, ... | | Authors: | Zhang, X, Zhan, W.Q, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Combating the SARS-CoV-2 Omicron (BA.1) and BA.2 with potent bispecific antibodies engineered from non-Omicron neutralizing antibodies
Cell Discov, 8, 2022
|
|
7WOP
 
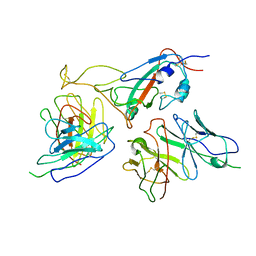 | | The local refined map of Omicron spike with bispecific antibody FD01 | | Descriptor: | 16L9, GW01, Spike protein S1 | | Authors: | Zhang, X, Zhan, W.Q, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Combating the SARS-CoV-2 Omicron (BA.1) and BA.2 with potent bispecific antibodies engineered from non-Omicron neutralizing antibodies
Cell Discov, 8, 2022
|
|
7WOR
 
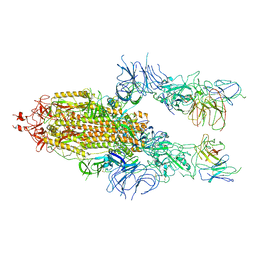 | | The state 2 of Omicron Spike with bispecific antibody FD01 | | Descriptor: | 16L9 Fv, 2-acetamido-2-deoxy-beta-D-glucopyranose, GW01 Fv, ... | | Authors: | Zhang, X, Zhan, W.Q, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Combating the SARS-CoV-2 Omicron (BA.1) and BA.2 with potent bispecific antibodies engineered from non-Omicron neutralizing antibodies
Cell Discov, 8, 2022
|
|
