3Q4J
 
 | | Structure of a small peptide ligand bound to E.coli DNA sliding clamp | | 分子名称: | DNA polymerase III subunit beta, peptide ligand | | 著者 | Wolff, P, Olieric, V, Briand, J.P, Chaloin, O, Dejaegere, A, Dumas, P, Ennifar, E, Guichard, G, Wagner, J, Burnouf, D. | | 登録日 | 2010-12-23 | | 公開日 | 2011-12-28 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure-based design of short peptide ligands binding onto the E. coli processivity ring.
J.Med.Chem., 54, 2011
|
|
3Q4L
 
 | | Structure of a small peptide ligand bound to E.coli DNA sliding clamp | | 分子名称: | DNA polymerase III subunit beta, SODIUM ION, peptide ligand | | 著者 | Wolff, P, Olieric, V, Briand, J.P, Chaloin, O, Dejaegere, A, Dumas, P, Ennifar, E, Guichard, G, Wagner, J, Burnouf, D. | | 登録日 | 2010-12-23 | | 公開日 | 2011-12-28 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure-based design of short peptide ligands binding onto the E. coli processivity ring.
J.Med.Chem., 54, 2011
|
|
3Q4K
 
 | | Structure of a small peptide ligand bound to E.coli DNA sliding clamp | | 分子名称: | DNA polymerase III subunit beta, peptide ligand | | 著者 | Wolff, P, Olieric, V, Briand, J.P, Chaloin, O, Dejaegere, A, Dumas, P, Ennifar, E, Guichard, G, Wagner, J, Burnouf, D. | | 登録日 | 2010-12-23 | | 公開日 | 2011-12-28 | | 最終更新日 | 2013-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure-based design of short peptide ligands binding onto the E. coli processivity ring.
J.Med.Chem., 54, 2011
|
|
4TR7
 
 | |
4TR8
 
 | | Crystal structure of DNA polymerase sliding clamp from Pseudomonas aeruginosa | | 分子名称: | DNA polymerase III subunit beta, SODIUM ION | | 著者 | Olieric, V, Burnouf, D, Ennifar, E, Wolff, P. | | 登録日 | 2014-06-15 | | 公開日 | 2014-09-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Differential Modes of Peptide Binding onto Replicative Sliding Clamps from Various Bacterial Origins.
J.Med.Chem., 57, 2014
|
|
4TR6
 
 | | Crystal structure of DNA polymerase sliding clamp from Bacillus subtilis | | 分子名称: | DNA polymerase III subunit beta, SODIUM ION | | 著者 | Burnouf, D, Olieric, V, Ennifar, E, Wolff, P. | | 登録日 | 2014-06-14 | | 公開日 | 2014-09-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Differential Modes of Peptide Binding onto Replicative Sliding Clamps from Various Bacterial Origins.
J.Med.Chem., 57, 2014
|
|
4TSZ
 
 | | Crystal structure of DNA polymerase sliding clamp from Pseudomonas aeruginosa with ligand | | 分子名称: | ACE-GLN-ALC-ASP-LEU-ZCL peptide, DNA polymerase III subunit beta | | 著者 | Olieric, V, Burnouf, D, Ennifar, E, Wolff, P. | | 登録日 | 2014-06-19 | | 公開日 | 2014-09-10 | | 最終更新日 | 2016-12-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Differential Modes of Peptide Binding onto Replicative Sliding Clamps from Various Bacterial Origins.
J.Med.Chem., 57, 2014
|
|
3ITH
 
 | | Crystal structure of the HIV-1 reverse transcriptase bound to a 6-vinylpyrimidine inhibitor | | 分子名称: | 6-ethenyl-N,N-dimethyl-2-(methylsulfonyl)pyrimidin-4-amine, Reverse transcriptase/ribonuclease H, p51 RT | | 著者 | Freisz, S, Bec, G, Wolff, P, Dumas, P, Radi, M, Botta, M. | | 登録日 | 2009-08-28 | | 公開日 | 2010-03-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal Structure of HIV-1 Reverse Transcriptase Bound to a Non-Nucleoside Inhibitor with a Novel Mechanism of Action
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
8PAY
 
 | | Structure of the E.coli DNA polymerase sliding clamp with a covalently bound peptide 2. | | 分子名称: | ACE-GLN-ALC-GLC-LEU-PHE, Beta sliding clamp, GLYCEROL, ... | | 著者 | Compain, G, Monsarrat, C, Blagojevic, J, Brillet, K, Dumas, P, Hammann, P, Kuhn, L, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Burnouf, D, wagner, J, Guichard, G. | | 登録日 | 2023-06-08 | | 公開日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.21 Å) | | 主引用文献 | Peptide-Based Covalent Inhibitors Bearing Mild Electrophiles to Target a Conserved His Residue of the Bacterial Sliding Clamp.
Jacs Au, 4, 2024
|
|
8PAT
 
 | | Structure of the E.coli DNA polymerase sliding clamp with a covalently bound peptide 3. | | 分子名称: | ACE-GLN-ALC-GLX-LEU-PHE, Beta sliding clamp | | 著者 | Compain, G, Monsarrat, C, Blagojevic, J, Brillet, K, Dumas, P, Hammann, P, Kuhn, L, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Burnouf, D, Guichard, G. | | 登録日 | 2023-06-08 | | 公開日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Peptide-Based Covalent Inhibitors Bearing Mild Electrophiles to Target a Conserved His Residue of the Bacterial Sliding Clamp.
Jacs Au, 4, 2024
|
|
1Q7X
 
 | | Solution structure of the alternatively spliced PDZ2 domain (PDZ2b) of PTP-Bas (hPTP1E) | | 分子名称: | PDZ2b domain of PTP-Bas (hPTP1E) | | 著者 | Kachel, N, Erdmann, K.S, Kremer, W, Wolff, P, Gronwald, W, Heumann, R, Kalbitzer, H.R, Structural Proteomics in Europe (SPINE) | | 登録日 | 2003-08-20 | | 公開日 | 2003-12-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure determination and ligand interactions of the PDZ2b domain of PTP-Bas (hPTP1E): Splicing induced modulation of ligand specificity.
J.Mol.Biol., 334, 2003
|
|
7AZ5
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 47 bound | | 分子名称: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, Peptide 47, ... | | 著者 | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | 登録日 | 2020-11-16 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZG
 
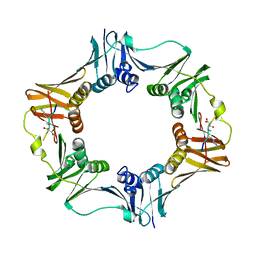 | | DNA polymerase sliding clamp from Escherichia coli with peptide 4 bound | | 分子名称: | Beta sliding clamp, Peptide 4 | | 著者 | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | 登録日 | 2020-11-16 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.92 Å) | | 主引用文献 | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZL
 
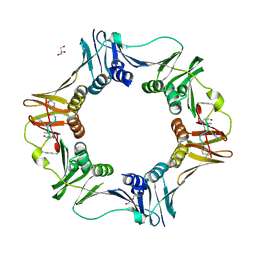 | | DNA polymerase sliding clamp from Escherichia coli with peptide 38 bound | | 分子名称: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | 登録日 | 2020-11-16 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZ7
 
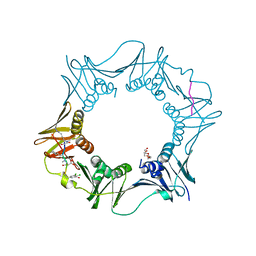 | | DNA polymerase sliding clamp from Escherichia coli with peptide 37 bound | | 分子名称: | Beta sliding clamp, FORMIC ACID, PENTAETHYLENE GLYCOL, ... | | 著者 | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | 登録日 | 2020-11-16 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZE
 
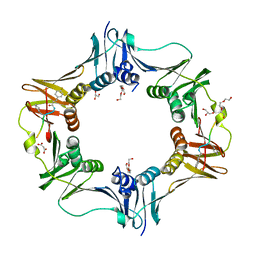 | | DNA polymerase sliding clamp from Escherichia coli with peptide 18 bound | | 分子名称: | Beta sliding clamp, GLYCEROL, MALONATE ION, ... | | 著者 | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | 登録日 | 2020-11-16 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZ6
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 36 bound | | 分子名称: | ACETATE ION, Beta sliding clamp, CHLORIDE ION, ... | | 著者 | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | 登録日 | 2020-11-16 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZ8
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 43 bound | | 分子名称: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | 登録日 | 2020-11-16 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZC
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 22 bound | | 分子名称: | Beta sliding clamp, GLYCEROL, Peptide 22 | | 著者 | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | 登録日 | 2020-11-16 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZF
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 8 bound | | 分子名称: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | 登録日 | 2020-11-16 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZD
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 20 bound | | 分子名称: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | 著者 | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | 登録日 | 2020-11-16 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZK
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 35 bound | | 分子名称: | Beta sliding clamp, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | 登録日 | 2020-11-16 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
8QOI
 
 | | Structure of the human 80S ribosome at 1.9 A resolution - the molecular role of chemical modifications and ions in RNA | | 分子名称: | 18S rRNA (1740-MER), 28S rRNA (3773-MER), 40S ribosomal protein S10, ... | | 著者 | Holvec, S, Barchet, C, Frechin, L, Hazemann, I, von Loeffelholz, O, Klaholz, B.P. | | 登録日 | 2023-09-29 | | 公開日 | 2024-06-12 | | 最終更新日 | 2024-08-28 | | 実験手法 | ELECTRON MICROSCOPY (1.9 Å) | | 主引用文献 | The structure of the human 80S ribosome at 1.9 angstrom resolution reveals the molecular role of chemical modifications and ions in RNA.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3ISN
 
 | | Crystal structure of HIV-1 RT bound to A 6-vinylpyrimidine inhibitor | | 分子名称: | 6-ethenyl-N,N-dimethyl-2-(methylsulfonyl)pyrimidin-4-amine, Reverse transcriptase/ribonuclease H, p51 RT | | 著者 | Ennifar, E, Freisz, S, Bec, G, Dumas, P, Botta, M, Radi, M. | | 登録日 | 2009-08-26 | | 公開日 | 2010-03-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of HIV-1 Reverse Transcriptase Bound to a Non-Nucleoside Inhibitor with a Novel Mechanism of Action
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
7NZ9
 
 | |
