8YSB
 
 | | Crystal structure of DynA1, a putative monoxygenase from Mivromonospora chersina. | | 分子名称: | Predicted ester cyclase | | 著者 | Yan, X.F, Huang, H.W, Gao, Y.G, Liang, Z.X. | | 登録日 | 2024-03-22 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-09-11 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | An Enzymatic Oxidation Cascade Converts delta-Thiolactone Anthracene to Anthraquinone in the Biosynthesis of Anthraquinone-Fused Enediynes.
Jacs Au, 4, 2024
|
|
8G0M
 
 | | Structure of complex between TV6.6 and CD98hc ECD | | 分子名称: | 1,2-ETHANEDIOL, 4F2 cell-surface antigen heavy chain, TETRAETHYLENE GLYCOL, ... | | 著者 | Kariolis, M.S, Lexa, K, Liau, N.P.D, Srivastava, D, Tran, H, Wells, R.C. | | 登録日 | 2023-01-31 | | 公開日 | 2023-10-18 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | CD98hc is a target for brain delivery of biotherapeutics.
Nat Commun, 14, 2023
|
|
8DMB
 
 | | Structure of Desulfovirgula thermocuniculi IsrB (DtIsrB) in complex with omega RNA and target DNA | | 分子名称: | MAGNESIUM ION, Ubiquitin-like protein SMT3,IsrB protein,monomeric superfolder Green Fluorescent Protein, non-target DNA, ... | | 著者 | Seiichi, H, Kappel, K, Zhang, F. | | 登録日 | 2022-07-08 | | 公開日 | 2022-10-19 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structure of the OMEGA nickase IsrB in complex with omega RNA and target DNA.
Nature, 610, 2022
|
|
8H1J
 
 | | Cryo-EM structure of the TnpB-omegaRNA-target DNA ternary complex | | 分子名称: | Non-target strand, RNA-guided DNA endonuclease TnpB, Target strand, ... | | 著者 | Nakagawa, R, Hirano, H, Omura, S, Nureki, O. | | 登録日 | 2022-10-03 | | 公開日 | 2023-04-12 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Cryo-EM structure of the transposon-associated TnpB enzyme.
Nature, 616, 2023
|
|
8W1P
 
 | |
8GKH
 
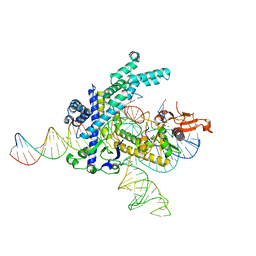 | | Structure of the Spizellomyces punctatus Fanzor (SpuFz) in complex with omega RNA and target DNA | | 分子名称: | DNA (35-MER), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), MAGNESIUM ION, ... | | 著者 | Xu, P, Saito, M, Zhang, F. | | 登録日 | 2023-03-19 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Fanzor is a eukaryotic programmable RNA-guided endonuclease.
Nature, 620, 2023
|
|
7VTI
 
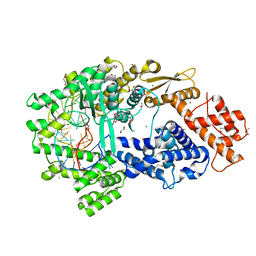 | | Crystal structure of the Cas13bt3-crRNA binary complex | | 分子名称: | 1,2-ETHANEDIOL, BROMIDE ION, CHLORIDE ION, ... | | 著者 | Nakagawa, R, Takeda, N.S, Tomita, A, Hirano, H, Kusakizako, T, Nishizawa, T, Yamashita, K, Nishimasu, H, Nureki, O. | | 登録日 | 2021-10-29 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Structure and engineering of the minimal type VI CRISPR-Cas13bt3.
Mol.Cell, 82, 2022
|
|
7VTN
 
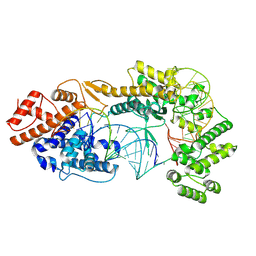 | | Cryo-EM structure of the Cas13bt3-crRNA-target RNA ternary complex | | 分子名称: | Cas13bt3, crRNA, target RNA | | 著者 | Nakagawa, R, Soumya, K, Han, A, Takeda, N.S, Tomita, A, Hirano, H, Kusakizako, T, Tomohiro, N, Yamashita, K, Feng, Z, Nishimasu, H, Nureki, O. | | 登録日 | 2021-10-30 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.38 Å) | | 主引用文献 | Structure and engineering of the minimal type VI CRISPR-Cas13bt3.
Mol.Cell, 82, 2022
|
|
7XHT
 
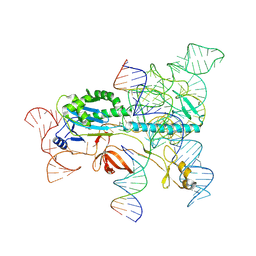 | | Structure of the OgeuIscB-omega RNA-target DNA complex | | 分子名称: | DNA (49-MER), DNA (5'-D(P*GP*AP*AP*GP*AP*AP*AP*AP*CP*CP*AP*T)-3'), LAURYL DIMETHYLAMINE-N-OXIDE, ... | | 著者 | Kato, K, Okazaki, O, Isayama, Y, Ishikawa, J, Nishizawa, T, Nishimasu, H. | | 登録日 | 2022-04-10 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.55 Å) | | 主引用文献 | Structure of the IscB-omega RNA ribonucleoprotein complex, the likely ancestor of CRISPR-Cas9.
Nat Commun, 13, 2022
|
|
3LMB
 
 | | The crystal structure of the protein OLEI01261 with unknown function from Chlorobaculum tepidum TLS | | 分子名称: | Uncharacterized protein | | 著者 | Zhang, R, Evdokimova, E, Egorova, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-01-29 | | 公開日 | 2010-03-16 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3LNP
 
 | | Crystal Structure of Amidohydrolase family Protein OLEI01672_1_465 from Oleispira antarctica | | 分子名称: | ACETIC ACID, Amidohydrolase family Protein OLEI01672_1_465, CALCIUM ION, ... | | 著者 | Kim, Y, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-02-02 | | 公開日 | 2010-02-16 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3I4Q
 
 | | Structure of a putative inorganic pyrophosphatase from the oil-degrading bacterium Oleispira antarctica | | 分子名称: | APC40078, SODIUM ION | | 著者 | Singer, A.U, Evdokimova, E, Kagan, O, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-07-02 | | 公開日 | 2009-07-28 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3M16
 
 | | Structure of a Transaldolase from Oleispira antarctica | | 分子名称: | Transaldolase | | 著者 | Singer, A.U, Kagan, O, Zhang, R, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-03-04 | | 公開日 | 2010-06-23 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3IRU
 
 | | Crystal structure of phoshonoacetaldehyde hydrolase like protein from Oleispira antarctica | | 分子名称: | SODIUM ION, phoshonoacetaldehyde hydrolase like protein | | 著者 | Chang, C, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-08-24 | | 公開日 | 2009-09-01 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
7ZR3
 
 | |
5WGT
 
 | | Crystal Structure of MalA' H253A, premalbrancheamide complex | | 分子名称: | (5aS,12aS,13aS)-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CADMIUM ION, CHLORIDE ION, ... | | 著者 | Fraley, A.E, Smith, J.L. | | 登録日 | 2017-07-14 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.093 Å) | | 主引用文献 | Function and Structure of MalA/MalA', Iterative Halogenases for Late-Stage C-H Functionalization of Indole Alkaloids.
J. Am. Chem. Soc., 139, 2017
|
|
5WGV
 
 | | Crystal Structure of MalA' C112S/C128S, premalbrancheamide complex | | 分子名称: | (5aS,12aS,13aS)-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CADMIUM ION, CHLORIDE ION, ... | | 著者 | Fraley, A.E, Smith, J.L. | | 登録日 | 2017-07-14 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Function and Structure of MalA/MalA', Iterative Halogenases for Late-Stage C-H Functionalization of Indole Alkaloids.
J. Am. Chem. Soc., 139, 2017
|
|
5WGY
 
 | | Crystal Structure of MalA' C112S/C128S, malbrancheamide B complex | | 分子名称: | (5aS,12aS,13aS)-9-chloro-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CADMIUM ION, CHLORIDE ION, ... | | 著者 | Fraley, A.E, Smith, J.L. | | 登録日 | 2017-07-14 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Function and Structure of MalA/MalA', Iterative Halogenases for Late-Stage C-H Functionalization of Indole Alkaloids.
J. Am. Chem. Soc., 139, 2017
|
|
5WGW
 
 | | Crystal Structure of Wild-type MalA', malbrancheamide B complex | | 分子名称: | (5aS,12aS,13aS)-9-chloro-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CADMIUM ION, CHLORIDE ION, ... | | 著者 | Fraley, A.E, Smith, J.L. | | 登録日 | 2017-07-14 | | 公開日 | 2017-08-16 | | 最終更新日 | 2019-11-27 | | 実験手法 | X-RAY DIFFRACTION (2.092 Å) | | 主引用文献 | Function and Structure of MalA/MalA', Iterative Halogenases for Late-Stage C-H Functionalization of Indole Alkaloids.
J. Am. Chem. Soc., 139, 2017
|
|
5WGU
 
 | | Crystal Structure of MalA' E494D, premalbrancheamide complex | | 分子名称: | (5aS,12aS,13aS)-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CADMIUM ION, CHLORIDE ION, ... | | 著者 | Fraley, A.E, Smith, J.L. | | 登録日 | 2017-07-14 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.052 Å) | | 主引用文献 | Function and Structure of MalA/MalA', Iterative Halogenases for Late-Stage C-H Functionalization of Indole Alkaloids.
J. Am. Chem. Soc., 139, 2017
|
|
5WGS
 
 | | Crystal Structure of MalA' H253F, premalbrancheamide complex | | 分子名称: | (5aS,12aS,13aS)-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CADMIUM ION, CHLORIDE ION, ... | | 著者 | Fraley, A.E, Smith, J.L. | | 登録日 | 2017-07-14 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Function and Structure of MalA/MalA', Iterative Halogenases for Late-Stage C-H Functionalization of Indole Alkaloids.
J. Am. Chem. Soc., 139, 2017
|
|
5WGR
 
 | | Crystal Structure of Wild-type MalA', premalbrancheamide complex | | 分子名称: | (5aS,12aS,13aS)-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CADMIUM ION, CHLORIDE ION, ... | | 著者 | Fraley, A.E, Smith, J.L. | | 登録日 | 2017-07-14 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.362 Å) | | 主引用文献 | Function and Structure of MalA/MalA', Iterative Halogenases for Late-Stage C-H Functionalization of Indole Alkaloids.
J. Am. Chem. Soc., 139, 2017
|
|
5WGZ
 
 | | Crystal Structure of Wild-type MalA', isomalbrancheamide B complex | | 分子名称: | (5aS,12aS,13aS)-8-chloro-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CADMIUM ION, CHLORIDE ION, ... | | 著者 | Fraley, A.E, Smith, J.L. | | 登録日 | 2017-07-14 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.041 Å) | | 主引用文献 | Function and Structure of MalA/MalA', Iterative Halogenases for Late-Stage C-H Functionalization of Indole Alkaloids.
J. Am. Chem. Soc., 139, 2017
|
|
5WGX
 
 | | Crystal Structure of MalA' H253A, malbrancheamide B complex | | 分子名称: | (5aS,12aS,13aS)-9-chloro-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CADMIUM ION, CHLORIDE ION, ... | | 著者 | Fraley, A.E, Smith, J.L. | | 登録日 | 2017-07-14 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.973 Å) | | 主引用文献 | Function and Structure of MalA/MalA', Iterative Halogenases for Late-Stage C-H Functionalization of Indole Alkaloids.
J. Am. Chem. Soc., 139, 2017
|
|
3LQY
 
 | | Crystal structure of putative isochorismatase hydrolase from Oleispira antarctica | | 分子名称: | GLYCEROL, putative isochorismatase hydrolase | | 著者 | Goral, A, Chruszcz, M, Kagan, O, Cymborowski, M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-02-10 | | 公開日 | 2010-03-16 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structure of a putative isochorismatase hydrolase from Oleispira antarctica.
J.Struct.Funct.Genom., 13, 2012
|
|
