1OBO
 
 | | W57L flavodoxin from Anabaena | | 分子名称: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN, SULFATE ION | | 著者 | Romero, A, Ramon, A, Fernandez-Cabrera, C, Irun, M.P, Sancho, J. | | 登録日 | 2003-01-31 | | 公開日 | 2003-04-24 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | How Fmn Binds to Anabaena Apoflavodoxin: A Hydrophobic Encounter at an Open Binding Site
J.Biol.Chem., 278, 2003
|
|
1OBV
 
 | | Y94F flavodoxin from Anabaena | | 分子名称: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN, SULFATE ION | | 著者 | Romero, A, Ramon, A, Fernandez-Cabrera, C, Irun, M.P, Sancho, J. | | 登録日 | 2003-01-31 | | 公開日 | 2003-04-24 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | How Fmn Binds to Anabaena Apoflavodoxin: A Hydrophobic Encounter at an Open Binding Site
J.Biol.Chem., 278, 2003
|
|
2XZA
 
 | | Crystal Structure of recombinant A.17 antibody FAB fragment | | 分子名称: | FAB A.17 HEAVY CHAIN, FAB A.17 LIGHT CHAIN | | 著者 | Carletti, E, Nachon, F, Nicolet, Y, Masson, P, Kurkova, I, Smirnov, I, Friboulet, A, Tramontano, A, Gabibov, A. | | 登録日 | 2010-11-24 | | 公開日 | 2011-09-21 | | 最終更新日 | 2020-03-11 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Reactibodies Generated by Kinetic Selection Couple Chemical Reactivity with Favorable Protein Dynamics.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2XZC
 
 | | Crystal Structure of phosphonate-modified recombinant A.17 antibody FAB fragment | | 分子名称: | 8-METHYL-8-AZABICYCLO[3.2.1]OCTAN-3-YL PHENYLPHOSPHONATE, CHLORIDE ION, FAB A.17 HEAVY CHAIN, ... | | 著者 | Carletti, E, Nachon, F, Nicolet, Y, Masson, P, Kurkova, I, Smirnov, I, Friboulet, A, Tramontano, A, Gabibov, A. | | 登録日 | 2010-11-24 | | 公開日 | 2011-09-21 | | 最終更新日 | 2020-03-11 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Reactibodies Generated by Kinetic Selection Couple Chemical Reactivity with Favorable Protein Dynamics.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7ZTH
 
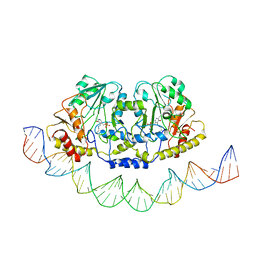 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the open conformation | | 分子名称: | DNA (48-MER), PLP-dependent aminotransferase family protein | | 著者 | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | 登録日 | 2022-05-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZLA
 
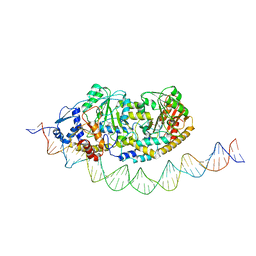 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the half-closed conformation | | 分子名称: | DNA (48-MER), PLP-dependent aminotransferase family protein | | 著者 | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Savino, C, Exertier, C, Bolognesi, M, Chaves Sanjuan, A. | | 登録日 | 2022-04-14 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (3.99 Å) | | 主引用文献 | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZN5
 
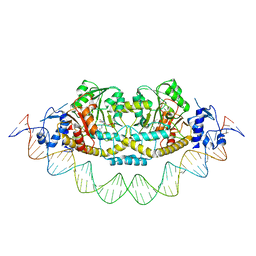 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C2 symmetry. | | 分子名称: | DNA (48-MER), PLP-dependent aminotransferase family protein | | 著者 | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | 登録日 | 2022-04-20 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZPA
 
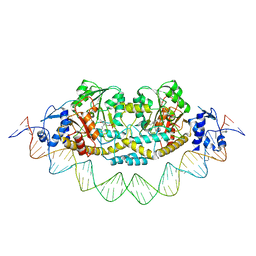 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C1 symmetry | | 分子名称: | DNA (48-MER), PLP-dependent aminotransferase family protein | | 著者 | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | 登録日 | 2022-04-27 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
6YLZ
 
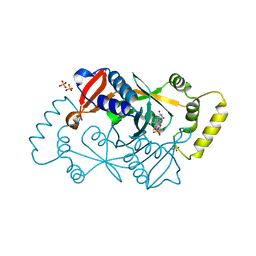 | | X-ray structure of the K72I,Y129F,R133L, H199A quadruple mutant of PNP-oxidase from E. coli | | 分子名称: | FLAVIN MONONUCLEOTIDE, PHOSPHATE ION, Pyridoxine/pyridoxamine 5'-phosphate oxidase, ... | | 著者 | Battista, T, Sularea, M, Barile, A, Fiorillo, A, Tramonti, A, Ilari, A. | | 登録日 | 2020-04-07 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.558 Å) | | 主引用文献 | Identification and characterization of the pyridoxal 5'-phosphate allosteric site in Escherichia coli pyridoxine 5'-phosphate oxidase.
J.Biol.Chem., 296, 2021
|
|
6YMH
 
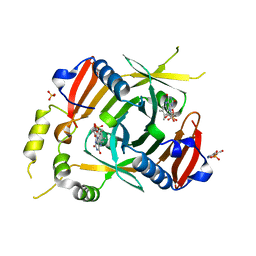 | | X-ray structure of the K72I, Y129F, R133L, H199A quadruple mutant of PNP-oxidase from E. coli in complex with PLP | | 分子名称: | FLAVIN MONONUCLEOTIDE, PYRIDOXAL-5'-PHOSPHATE, Pyridoxine/pyridoxamine 5'-phosphate oxidase, ... | | 著者 | Battista, T, Sularea, M, Barile, A, Fiorillo, A, Tramonti, A, Ilari, A. | | 登録日 | 2020-04-08 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.417 Å) | | 主引用文献 | Identification and characterization of the pyridoxal 5'-phosphate allosteric site in Escherichia coli pyridoxine 5'-phosphate oxidase.
J.Biol.Chem., 296, 2021
|
|
6YMF
 
 | | Crystal structure of serine hydroxymethyltransferase from Aphanothece halophytica in the PLP-Serine external aldimine state | | 分子名称: | GLYCEROL, PENTAETHYLENE GLYCOL, Serine hydroxymethyltransferase, ... | | 著者 | Ruszkowski, M, Sekula, B, Nogues, I, Tramonti, A, Angelaccio, S, Contestabile, R. | | 登録日 | 2020-04-08 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Structural and kinetic properties of serine hydroxymethyltransferase from the halophytic cyanobacterium Aphanothece halophytica provide a rationale for salt tolerance.
Int.J.Biol.Macromol., 159, 2020
|
|
6YMD
 
 | | Crystal structure of serine hydroxymethyltransferase from Aphanothece halophytica in the covalent complex with malonate | | 分子名称: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, MALONATE ION, ... | | 著者 | Ruszkowski, M, Sekula, B, Nogues, I, Tramonti, A, Angelaccio, S, Contestabile, R. | | 登録日 | 2020-04-08 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Structural and kinetic properties of serine hydroxymethyltransferase from the halophytic cyanobacterium Aphanothece halophytica provide a rationale for salt tolerance.
Int.J.Biol.Macromol., 159, 2020
|
|
6YME
 
 | | Crystal structure of serine hydroxymethyltransferase from Aphanothece halophytica in the PLP-internal aldimine state | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Serine hydroxymethyltransferase | | 著者 | Ruszkowski, M, Sekula, B, Nogues, I, Tramonti, A, Angelaccio, S, Contestabile, R. | | 登録日 | 2020-04-08 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Structural and kinetic properties of serine hydroxymethyltransferase from the halophytic cyanobacterium Aphanothece halophytica provide a rationale for salt tolerance.
Int.J.Biol.Macromol., 159, 2020
|
|
7UAX
 
 | | The crystal structure of the K36A/K38A double mutant of E. coli YGGS in complex with PLP | | 分子名称: | PHOSPHATE ION, Pyridoxal phosphate homeostasis protein | | 著者 | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | 登録日 | 2022-03-14 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7U9C
 
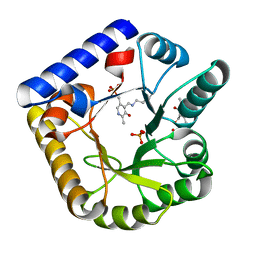 | | Crystal Structure of the wild type Escherichia coli Pyridoxal 5'-phosphate homeostasis protein (YGGS) | | 分子名称: | PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein | | 著者 | Donkor, A.K, Ghatge, M.S, Safo, M.K, Musayev, F.N. | | 登録日 | 2022-03-10 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7U9H
 
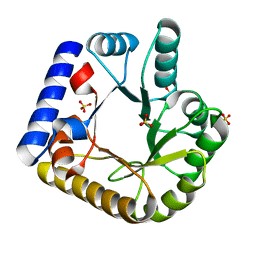 | | Crystal Structure of Escherichia coli apo Pyridoxal 5'-phosphate homeostasis protein (YGGS) | | 分子名称: | Pyridoxal phosphate homeostasis protein, SULFATE ION | | 著者 | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | 登録日 | 2022-03-10 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7UAT
 
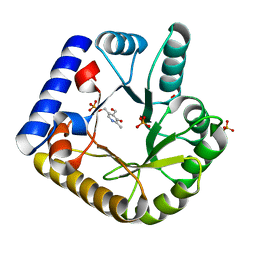 | | The crystal structure of the K36A mutant of E. coli YGGS in complex with PLP | | 分子名称: | PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein | | 著者 | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | 登録日 | 2022-03-14 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7UAU
 
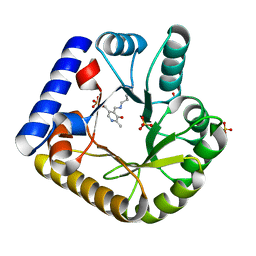 | | The crystal structure of the K137A mutant of E. coli YGGS in complex with PLP | | 分子名称: | PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein, SULFATE ION | | 著者 | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | 登録日 | 2022-03-14 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7UB8
 
 | | The crystal structure of the K38A/K137A/K233A/K234A quadruple mutant of E. coli YGGS in complex with PLP | | 分子名称: | 1,4-BUTANEDIOL, PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein | | 著者 | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | 登録日 | 2022-03-14 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7UBP
 
 | | The crystal structure of the K36A/K137A double mutant of E. coli YGGS in complex with PLP | | 分子名称: | PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein, SULFATE ION | | 著者 | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | 登録日 | 2022-03-15 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7UBQ
 
 | | The crystal structure of the wild-type of E. coli YGGS in complex with PNP | | 分子名称: | PYRIDOXINE-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein | | 著者 | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | 登録日 | 2022-03-15 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7UB4
 
 | | The crystal structure of the K36A/K38A/K233A/K234A quadruple mutant of E. coli YGGS in complex with PLP | | 分子名称: | PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein | | 著者 | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | 登録日 | 2022-03-14 | | 公開日 | 2022-03-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
8A11
 
 | | Cryo-EM structure of the Human SHMT1-RNA complex | | 分子名称: | PYRIDOXAL-5'-PHOSPHATE, Serine hydroxymethyltransferase, cytosolic | | 著者 | Spizzichino, S, Marabelli, C, Bharadwaj, A, Jakobi, A.J, Chaves-Sanjuan, A, Giardina, G, Bolognesi, M, Cutruzzola, F. | | 登録日 | 2022-05-30 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Structure-based mechanism of riboregulation of the metabolic enzyme SHMT1.
Mol.Cell, 2024
|
|
8R7H
 
 | | Cryo-EM structure of Human SHMT1 | | 分子名称: | Serine hydroxymethyltransferase, cytosolic | | 著者 | Spizzichino, S, Marabelli, C, Bharadwaj, A, Jakobi, A.J, Chaves-Sanjuan, A, Giardina, G, Bolognesi, M, Cutruzzola, F. | | 登録日 | 2023-11-24 | | 公開日 | 2024-07-24 | | 最終更新日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Structure-based mechanism of riboregulation of the metabolic enzyme SHMT1.
Mol.Cell, 84, 2024
|
|
8QYT
 
 | | Human Pyridoxine-5'-phosphate oxidase in complex with PLP | | 分子名称: | BETA-MERCAPTOETHANOL, FLAVIN MONONUCLEOTIDE, PYRIDOXAL-5'-PHOSPHATE, ... | | 著者 | Antonelli, L, Ilari, A, Fiorillo, A. | | 登録日 | 2023-10-26 | | 公開日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Identification of the pyridoxal 5'-phosphate allosteric site in human pyridox(am)ine 5'-phosphate oxidase.
Protein Sci., 33, 2024
|
|
