9LOT
 
 | | Crystal structure of Escherichia coli trptophanyl-tRNA synthetase in complex with N-piperidine ibrutinib | | 分子名称: | 3-(4-phenoxyphenyl)-1-piperidin-4-yl-pyrazolo[3,4-d]pyrimidin-4-amine, CHLORZOXAZONE, SULFATE ION, ... | | 著者 | Peng, X, Chen, B, Xia, K, Zhou, H. | | 登録日 | 2025-01-23 | | 公開日 | 2025-05-07 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | The lineage-specific tRNA recognition mechanism of bacterial trptophanyl-tRNA synthetase and its implications for inhibitor discover
To Be Published
|
|
9LPD
 
 | | Crystal structure of Escherichia coli trptophanyl-tRNA synthetase in complex with tirabrutinib | | 分子名称: | CHLORZOXAZONE, SULFATE ION, TRYPTOPHANYL-5'AMP, ... | | 著者 | Peng, X, Chen, B, Xia, K, Zhou, H. | | 登録日 | 2025-01-24 | | 公開日 | 2025-05-07 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | The lineage-specific tRNA recognition mechanism of bacterial trptophanyl-tRNA synthetase and its implications for inhibitor discover
To Be Published
|
|
9LPC
 
 | |
7F61
 
 | |
8HBA
 
 | |
8HB1
 
 | | Crystal structure of NAD-II riboswitch (two strands) with NMN | | 分子名称: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, MAGNESIUM ION, RNA (30-MER), ... | | 著者 | Peng, X, Lilley, D.M.J, Huang, L. | | 登録日 | 2022-10-27 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Crystal structures of the NAD+-II riboswitch reveal two distinct ligand-binding pockets.
Nucleic Acids Res., 51, 2023
|
|
8HB8
 
 | |
8HB3
 
 | |
8I3Z
 
 | | Crystal structure of NAD-II riboswitch (two strands) with NMN at 1.67 angstrom | | 分子名称: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, RNA (31-MER), RNA (5'-R(*AP*GP*AP*GP*CP*GP*UP*UP*GP*CP*GP*UP*CP*CP*GP*AP*AP*AP*GP*UP*(CBV)P*GP*CP*C)-3'), ... | | 著者 | Peng, X, Lilley, D.M.J, Huang, L. | | 登録日 | 2023-01-18 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Crystal structures of the NAD+-II riboswitch reveal two distinct ligand-binding pockets.
Nucleic Acids Res., 51, 2023
|
|
8K85
 
 | | Crystal structure of Red Broccoli aptamer with OBI | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, (5Z)-3-(3H-benzimidazol-4-ylmethyl)-5-[[3,5-bis(fluoranyl)-4-oxidanyl-phenyl]methylidene]-2-[(E)-hydroxyiminomethyl]imidazol-4-one, POTASSIUM ION, ... | | 著者 | Peng, X, Huang, L. | | 登録日 | 2023-07-28 | | 公開日 | 2025-02-12 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Structural analysis of various Broccoli aptamers with different Fluorophores
To Be Published
|
|
3EFH
 
 | |
8K7W
 
 | | Crystal structure of Broccoli aptamer with DFHBI-1T | | 分子名称: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, POTASSIUM ION, RNA (49-MER), ... | | 著者 | Peng, X, Huang, L. | | 登録日 | 2023-07-27 | | 公開日 | 2025-01-29 | | 最終更新日 | 2025-03-19 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | A general strategy for engineering GU base pairs to facilitate RNA crystallization.
Nucleic Acids Res., 53, 2025
|
|
9L0R
 
 | |
7E27
 
 | | Structure of PfFNT in complex with MMV007839 | | 分子名称: | (Z)-4,4,5,5,5-pentakis(fluoranyl)-1-(4-methoxy-2-oxidanyl-phenyl)-3-oxidanyl-pent-2-en-1-one, Formate-nitrite transporter | | 著者 | Yan, C.Y, Jiang, X, Deng, D, Peng, X, Wang, N, Zhu, A, Xu, H, Li, J. | | 登録日 | 2021-02-04 | | 公開日 | 2021-08-18 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.29 Å) | | 主引用文献 | Structural characterization of the Plasmodium falciparum lactate transporter PfFNT alone and in complex with antimalarial compound MMV007839 reveals its inhibition mechanism.
Plos Biol., 19, 2021
|
|
7E26
 
 | | Structure of PfFNT in apo state | | 分子名称: | Formate-nitrite transporter | | 著者 | Yan, C.Y, Jiang, X, Deng, D, Peng, X, Wang, N, Zhu, A, Xu, H, Li, J. | | 登録日 | 2021-02-04 | | 公開日 | 2021-08-18 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.29 Å) | | 主引用文献 | Structural characterization of the Plasmodium falciparum lactate transporter PfFNT alone and in complex with antimalarial compound MMV007839 reveals its inhibition mechanism.
Plos Biol., 19, 2021
|
|
1Z9M
 
 | | Crystal Structure of Nectin-like molecule-1 protein Domain 1 | | 分子名称: | GAPA225 | | 著者 | Dong, X, Xu, F, Gong, Y, Gao, J, Lin, P, Chen, T, Peng, Y, Qiang, B, Yuan, J, Peng, X, Rao, Z. | | 登録日 | 2005-04-03 | | 公開日 | 2006-02-07 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure of the V Domain of Human Nectin-like Molecule-1/Syncam3/Tsll1/Igsf4b, a Neural Tissue-specific Immunoglobulin-like Cell-Cell Adhesion Molecule
J.Biol.Chem., 281, 2006
|
|
9LMF
 
 | |
1DGI
 
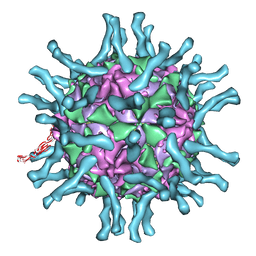 | | Cryo-EM structure of human poliovirus(serotype 1)complexed with three domain CD155 | | 分子名称: | POLIOVIRUS RECEPTOR, VP1, VP2, ... | | 著者 | He, Y, Bowman, V.D, Mueller, S, Bator, C.M, Bella, J, Peng, X, Baker, T.S, Wimmer, E, Kuhn, R.J, Rossmann, M.G. | | 登録日 | 1999-11-24 | | 公開日 | 2000-01-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | ELECTRON MICROSCOPY (22 Å) | | 主引用文献 | Interaction of the poliovirus receptor with poliovirus.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1NN8
 
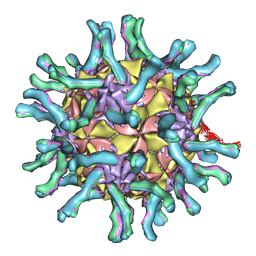 | | CryoEM structure of poliovirus receptor bound to poliovirus | | 分子名称: | MYRISTIC ACID, coat protein VP1, coat protein VP2, ... | | 著者 | He, Y, Mueller, S, Chipman, P.R, Bator, C.M, Peng, X, Bowman, V.D, Mukhopadhyay, S, Wimmer, E, Kuhn, R.J, Rossmann, M.G. | | 登録日 | 2003-01-13 | | 公開日 | 2004-01-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (15 Å) | | 主引用文献 | Complexes of poliovirus serotypes with their common cellular receptor, CD155
J.Virol., 77, 2003
|
|
6EXP
 
 | | Crystal structure of the SIRV3 AcrID1 (gp02) anti-CRISPR protein | | 分子名称: | SIRV3 AcrID1 (gp02) anti-CRISPR protein | | 著者 | He, F, Bhoobalan-Chitty, Y, Van, L.B, Kjeldsen, A.L, Dedola, M, Makarova, K.S, Koonin, E.V, Brodersen, D.E, Peng, X. | | 登録日 | 2017-11-08 | | 公開日 | 2018-01-31 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Anti-CRISPR proteins encoded by archaeal lytic viruses inhibit subtype I-D immunity.
Nat Microbiol, 3, 2018
|
|
4GHT
 
 | | Crystal structure of EV71 3C proteinase in complex with AG7088 | | 分子名称: | 3C proteinase, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | 著者 | Chen, C, Wu, C, Cai, Q, Li, N, Peng, X, Cai, Y, Yin, K, Chen, X, Wang, X, Zhang, R, Liu, L, Chen, S, Li, J, Lin, T. | | 登録日 | 2012-08-08 | | 公開日 | 2013-06-26 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Structures of Enterovirus 71 3C proteinase (strain E2004104-TW-CDC) and its complex with rupintrivir
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GHQ
 
 | | Crystal structure of EV71 3C proteinase | | 分子名称: | 3C proteinase | | 著者 | Chen, C, Wu, C, Cai, Q, Li, N, Peng, X, Cai, Y, Yin, K, Chen, X, Wang, X, Zhang, R, Liu, L, Chen, S, Li, J, Lin, T. | | 登録日 | 2012-08-08 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structures of Enterovirus 71 3C proteinase (strain E2004104-TW-CDC) and its complex with rupintrivir
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5DP4
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 3 | | 分子名称: | 3C proteinase, ethyl (2Z,4S)-4-{[(2S)-2-methyl-3-phenylpropanoyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | 著者 | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | 登録日 | 2015-09-12 | | 公開日 | 2016-03-30 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DP9
 
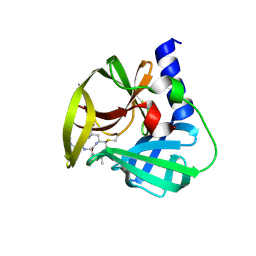 | | Crystal Structure of EV71 3C Proteinase in complex with compound 9 | | 分子名称: | 3C proteinase, ethyl (2Z,4S)-4-[(N-{[(cyclobutylmethyl)amino](oxo)acetyl}-L-phenylalanyl)amino]-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | 著者 | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | 登録日 | 2015-09-12 | | 公開日 | 2016-03-30 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DP6
 
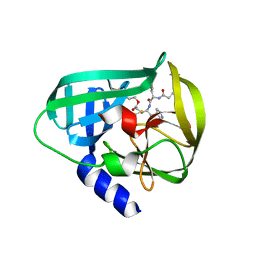 | | Crystal Structure of EV71 3C Proteinase in complex with compound 7 | | 分子名称: | 3C proteinase, ethyl (2Z,4S)-4-{[N-(3-cyclopropylpropanoyl)-L-phenylalanyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | 著者 | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | 登録日 | 2015-09-12 | | 公開日 | 2016-03-30 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
