6X6C
 
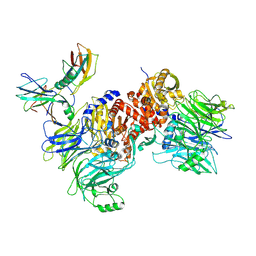 | | Cryo-EM structure of NLRP1-DPP9-VbP complex | | 分子名称: | Dipeptidyl peptidase 9, NACHT, LRR and PYD domains-containing protein 1, ... | | 著者 | Hollingsworth, L.R, Sharif, H, Griswold, A.R, Fontana, P, Mintseris, J, Dagbay, K.B, Paulo, J.A, Gygi, S.P, Bachovchin, D.A, Wu, H. | | 登録日 | 2020-05-27 | | 公開日 | 2021-03-10 | | 最終更新日 | 2021-05-12 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | DPP9 sequesters the C terminus of NLRP1 to repress inflammasome activation.
Nature, 592, 2021
|
|
6X6A
 
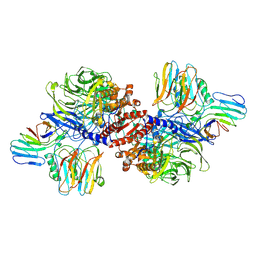 | | Cryo-EM structure of NLRP1-DPP9 complex | | 分子名称: | Dipeptidyl peptidase 9, NACHT, LRR and PYD domains-containing protein 1 | | 著者 | Hollingsworth, L.R, Sharif, H, Griswold, A.R, Fontana, P, Mintseris, J, Dagbay, K.B, Paulo, J.A, Gygi, S.P, Bachovchin, D.A, Wu, H. | | 登録日 | 2020-05-27 | | 公開日 | 2021-03-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | DPP9 sequesters the C terminus of NLRP1 to repress inflammasome activation.
Nature, 592, 2021
|
|
6BOF
 
 | |
7TMW
 
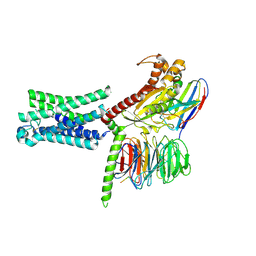 | | Cryo-EM structure of the relaxin receptor RXFP1 in complex with heterotrimeric Gs | | 分子名称: | Camelid antibody VHH fragment Nb35, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Erlandson, S.C, Rawson, S, Kruse, A.C. | | 登録日 | 2022-01-20 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-08-09 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | The relaxin receptor RXFP1 signals through a mechanism of autoinhibition.
Nat.Chem.Biol., 19, 2023
|
|
6MA3
 
 | | Crystal structure of human O-GlcNAc transferase bound to a peptide from HCF-1 pro-repeat 2 (11-26) and inhibitor 2a | | 分子名称: | 4-{2-[(1R)-2-{(carboxymethyl)[(thiophen-2-yl)methyl]amino}-2-oxo-1-{[(2-oxo-1,2-dihydroquinolin-6-yl)sulfonyl]amino}ethyl]phenoxy}butanoic acid, Host Cell Factor 1 peptide, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | 著者 | Martin, S.E.S, Lazarus, M.B, Walker, S. | | 登録日 | 2018-08-25 | | 公開日 | 2018-10-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-Based Evolution of Low Nanomolar O-GlcNAc Transferase Inhibitors.
J. Am. Chem. Soc., 140, 2018
|
|
6MA2
 
 | | Crystal structure of human O-GlcNAc transferase bound to a peptide from HCF-1 pro-repeat 2 (11-26) and inhibitor ent-1a | | 分子名称: | Host Cell Factor 1 peptide, N-[(2S)-2-(2-methoxyphenyl)-2-{[(2-oxo-1,2-dihydroquinolin-6-yl)sulfonyl]amino}acetyl]-N-[(thiophen-2-yl)methyl]glycine, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | 著者 | Martin, S.E.S, Lazarus, M.B, Walker, S. | | 登録日 | 2018-08-25 | | 公開日 | 2018-10-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-Based Evolution of Low Nanomolar O-GlcNAc Transferase Inhibitors.
J. Am. Chem. Soc., 140, 2018
|
|
6MA4
 
 | | Crystal structure of human O-GlcNAc transferase bound to a peptide from HCF-1 pro-repeat 2 (11-26) and inhibitor 3a | | 分子名称: | 5-{2-[(1R)-2-{(carboxymethyl)[(thiophen-2-yl)methyl]amino}-2-oxo-1-{[(2-oxo-1,2-dihydroquinolin-6-yl)sulfonyl]amino}ethyl]phenoxy}pentanoic acid, Host Cell Factor 1 peptide, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | 著者 | Martin, S.E.S, Lazarus, M.B, Walker, S. | | 登録日 | 2018-08-25 | | 公開日 | 2018-10-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-Based Evolution of Low Nanomolar O-GlcNAc Transferase Inhibitors.
J. Am. Chem. Soc., 140, 2018
|
|
6MA5
 
 | | Crystal structure of human O-GlcNAc transferase bound to a peptide from HCF-1 pro-repeat 2 (11-26) and inhibitor 1a | | 分子名称: | Host Cell Factor 1 peptide, N-[(2R)-2-(2-methoxyphenyl)-2-{[(2-oxo-1,2-dihydroquinolin-6-yl)sulfonyl]amino}acetyl]-N-[(thiophen-2-yl)methyl]glycine, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | 著者 | Martin, S.E.S, Lazarus, M.B, Walker, S. | | 登録日 | 2018-08-25 | | 公開日 | 2018-10-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-Based Evolution of Low Nanomolar O-GlcNAc Transferase Inhibitors.
J. Am. Chem. Soc., 140, 2018
|
|
6MA1
 
 | | Crystal structure of human O-GlcNAc transferase bound to a peptide from HCF-1 pro-repeat 2 (11-26) and inhibitor 4a | | 分子名称: | Host Cell Factor 1 peptide, N-[(2R)-2-{[(7-chloro-2-oxo-1,2-dihydroquinolin-6-yl)sulfonyl]amino}-2-(2-methoxyphenyl)acetyl]-N-[(thiophen-2-yl)methyl]glycine, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | 著者 | Martin, S.E.S, Lazarus, M.B, Walker, S. | | 登録日 | 2018-08-25 | | 公開日 | 2018-10-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structure-Based Evolution of Low Nanomolar O-GlcNAc Transferase Inhibitors.
J. Am. Chem. Soc., 140, 2018
|
|
6ZVH
 
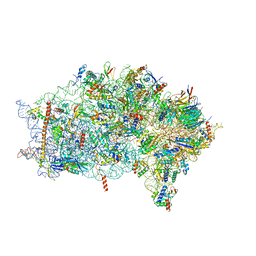 | | EDF1-ribosome complex | | 分子名称: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | 著者 | Best, K.M, Denk, T, Cheng, J, Thoms, M, Berninghausen, O, Beckmann, R. | | 登録日 | 2020-07-24 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | EDF1 coordinates cellular responses to ribosome collisions.
Elife, 9, 2020
|
|
6ZVI
 
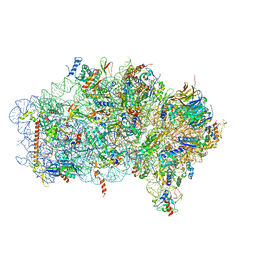 | | Mbf1-ribosome complex | | 分子名称: | 18S rRNA, 40S ribosomal protein S0-A, 40S ribosomal protein S10-A, ... | | 著者 | Best, K.M, Denk, T, Cheng, J, Thoms, M, Berninghausen, O, Beckmann, R. | | 登録日 | 2020-07-24 | | 公開日 | 2020-09-09 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | EDF1 coordinates cellular responses to ribosome collisions.
Elife, 9, 2020
|
|
8OJ5
 
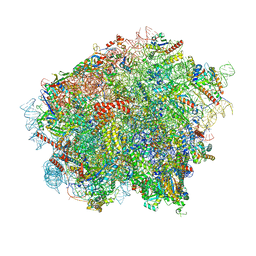 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 3 (in-vitro reconstitution) | | 分子名称: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Penchev, I, DaRosa, P.A, Peter, J.J, Kulathu, Y, Becker, T, Beckmann, R, Kopito, R. | | 登録日 | 2023-03-23 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
8OJ8
 
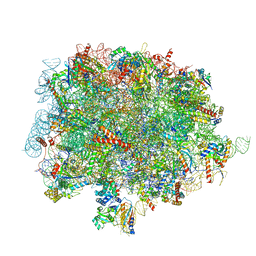 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 1 (native) | | 分子名称: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Penchev, I, DaRosa, P.A, Becker, T, Beckmann, R, Kopito, R. | | 登録日 | 2023-03-24 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
8OJ0
 
 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 2 (native) | | 分子名称: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Penchev, I, DaRosa, P.A, Becker, T, Beckmann, R, Kopito, R. | | 登録日 | 2023-03-23 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
8OHD
 
 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 3 (native) | | 分子名称: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Penchev, I, DaRosa, P.A, Becker, T, Beckmann, R, Kopito, R. | | 登録日 | 2023-03-21 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
6BP4
 
 | |
6BOX
 
 | |
5V83
 
 | | Structure of DCN1 bound to NAcM-HIT | | 分子名称: | Lysozyme,DCN1-like protein 1 chimera, N-(1-benzylpiperidin-4-yl)-N'-[3-(trifluoromethyl)phenyl]urea | | 著者 | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | 登録日 | 2017-03-21 | | 公開日 | 2017-05-24 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.002 Å) | | 主引用文献 | Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase.
Nat. Chem. Biol., 13, 2017
|
|
5V86
 
 | | Structure of DCN1 bound to NAcM-OPT | | 分子名称: | Lysozyme,DCN1-like protein 1, N-benzyl-N-(1-butylpiperidin-4-yl)-N'-(3,4-dichlorophenyl)urea | | 著者 | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | 登録日 | 2017-03-21 | | 公開日 | 2017-05-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.374 Å) | | 主引用文献 | Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase.
Nat. Chem. Biol., 13, 2017
|
|
5V88
 
 | | Structure of DCN1 bound to NAcM-COV | | 分子名称: | Lysozyme,DCN1-like protein 1, N-{2-[({1-[(2R)-pentan-2-yl]piperidin-4-yl}{[3-(trifluoromethyl)phenyl]carbamoyl}amino)methyl]phenyl}propanamide | | 著者 | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | 登録日 | 2017-03-21 | | 公開日 | 2017-05-24 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.601 Å) | | 主引用文献 | Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase.
Nat. Chem. Biol., 13, 2017
|
|
5V89
 
 | | Structure of DCN4 PONY domain bound to CUL1 WHB | | 分子名称: | Cullin-1, DCN1-like protein 4 | | 著者 | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | 登録日 | 2017-03-21 | | 公開日 | 2017-05-24 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase.
Nat. Chem. Biol., 13, 2017
|
|
7T9X
 
 | |
7T92
 
 | |
7JII
 
 | | HRAS A59E GDP | | 分子名称: | CALCIUM ION, GTPase HRas, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Johnson, C.W, Haigis, K.M. | | 登録日 | 2020-07-23 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.532 Å) | | 主引用文献 | Regulation of GTPase function by autophosphorylation.
Mol.Cell, 82, 2022
|
|
7JIF
 
 | | HRAS A59T GppNHp | | 分子名称: | GLYCEROL, GTPase HRas, MAGNESIUM ION, ... | | 著者 | Johnson, C.W, Haigis, K.M. | | 登録日 | 2020-07-23 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.757 Å) | | 主引用文献 | Regulation of GTPase function by autophosphorylation.
Mol.Cell, 82, 2022
|
|
