3FN1
 
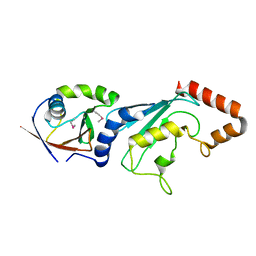 | | E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. | | 分子名称: | NEDD8-activating enzyme E1 catalytic subunit, NEDD8-conjugating enzyme UBE2F | | 著者 | Huang, D.T, Ayrault, O, Hunt, H.W, Taherbhoy, A.M, Duda, D.M, Scott, D.C, Borg, L.A, Neale, G, Murray, P.J, Roussel, M.F, Schulman, B.A. | | 登録日 | 2008-12-22 | | 公開日 | 2009-03-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification
Mol.Cell, 33, 2009
|
|
8PD0
 
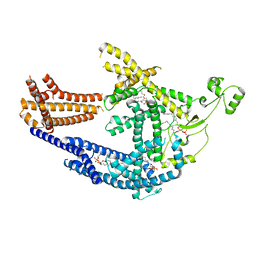 | | cryo-EM structure of Doa10 in MSP1E3D1 | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, ERAD-associated E3 ubiquitin-protein ligase DOA10 | | 著者 | Botsch, J.J, Braeuning, B, Schulman, B.A. | | 登録日 | 2023-06-11 | | 公開日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (3.58 Å) | | 主引用文献 | Doa10/MARCH6 architecture interconnects E3 ligase activity with lipid-binding transmembrane channel to regulate SQLE.
Nat Commun, 15, 2024
|
|
8PDA
 
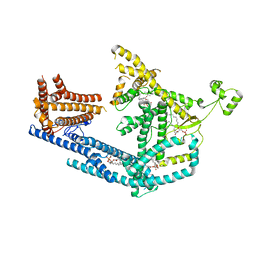 | | cryo-EM structure of Doa10 with RING domain in MSP1E3D1 | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, ERAD-associated E3 ubiquitin-protein ligase DOA10 | | 著者 | Botsch, J.J, Braeuning, B, Schulman, B.A. | | 登録日 | 2023-06-12 | | 公開日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (3.58 Å) | | 主引用文献 | Doa10/MARCH6 architecture interconnects E3 ligase activity with lipid-binding transmembrane channel to regulate SQLE.
Nat Commun, 15, 2024
|
|
8QBN
 
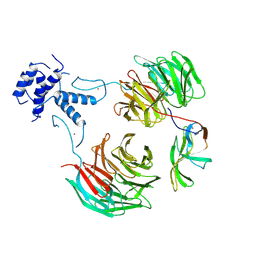 | |
8QE8
 
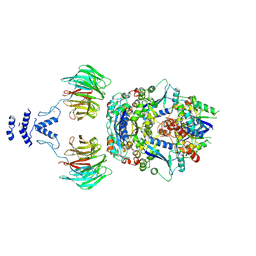 | | Structure of the non-canonical CTLH E3 substrate receptor WDR26 bound to NMNAT1 substrate | | 分子名称: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Nicotinamide/nicotinic acid mononucleotide adenylyltransferase 1, WD repeat-containing protein 26, ... | | 著者 | Chrustowicz, J, Sherpa, D, Schulman, B.A. | | 登録日 | 2023-08-30 | | 公開日 | 2024-05-15 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Non-canonical substrate recognition by the human WDR26-CTLH E3 ligase regulates prodrug metabolism.
Mol.Cell, 84, 2024
|
|
8CAF
 
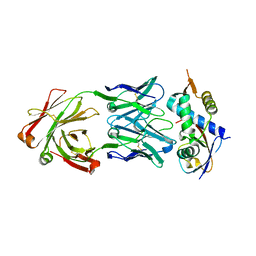 | | N8C_Fab3b in complex with NEDD8-CUL1(WHB) | | 分子名称: | Cullin-1, Fab Heavy Chain, Fab Light Chain, ... | | 著者 | Duda, D.M, Yanishevski, D, Henneberg, L.T, Schulman, B.A. | | 登録日 | 2023-01-24 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | Activity-based profiling of cullin-RING E3 networks by conformation-specific probes.
Nat.Chem.Biol., 19, 2023
|
|
6SYT
 
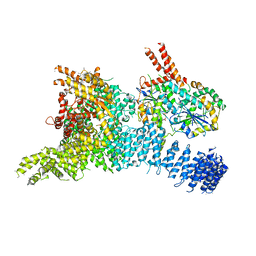 | | Structure of the SMG1-SMG8-SMG9 complex | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Gat, Y, Schuller, J.M, Conti, E. | | 登録日 | 2019-10-01 | | 公開日 | 2019-12-11 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.45 Å) | | 主引用文献 | InsP6binding to PIKK kinases revealed by the cryo-EM structure of an SMG1-SMG8-SMG9 complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
