8BA5
 
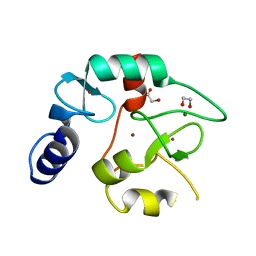 | | Crystal structure of the DNMT3A ADD domain | | 分子名称: | 1,2-ETHANEDIOL, DNA (cytosine-5)-methyltransferase 3A, MAGNESIUM ION, ... | | 著者 | Linhard, V, Kunert, S, Wollenhaupt, J, Broehm, A, Schwalbe, H, Jeltsch, A. | | 登録日 | 2022-10-11 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | The MECP2-TRD domain interacts with the DNMT3A-ADD domain at the H3-tail binding site.
Protein Sci., 32, 2023
|
|
1H56
 
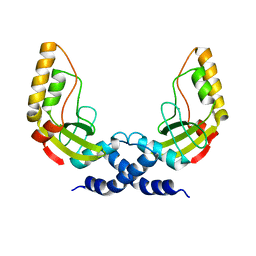 | | Structural and biochemical characterization of a new magnesium ion binding site near Tyr94 in the restriction endonuclease PvuII | | 分子名称: | MAGNESIUM ION, TYPE II RESTRICTION ENZYME PVUII | | 著者 | Spyrida, A, Matzen, C, Lanio, T, Jeltsch, A, Simoncsits, A, Athanasiadis, A, Scheuring-Vanamee, E, Kokkinidis, M, Pingoud, A. | | 登録日 | 2001-05-20 | | 公開日 | 2003-08-07 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural and Biochemical Characterization of a New Mg(2+) Binding Site Near Tyr94 in the Restriction Endonuclease PvuII.
J.Mol.Biol., 331, 2003
|
|
2G1P
 
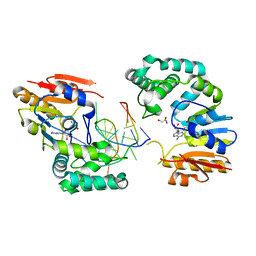 | | Structure of E. coli DNA adenine methyltransferase (DAM) | | 分子名称: | 5'-D(*TP*CP*TP*AP*GP*AP*TP*CP*TP*AP*GP*A)-3', DNA adenine methylase, GLYCEROL, ... | | 著者 | Horton, J.R, Liebert, K, Bekes, M, Jeltsch, A, Cheng, X. | | 登録日 | 2006-02-14 | | 公開日 | 2006-11-21 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Structure and substrate recognition of the Escherichia coli DNA adenine methyltransferase.
J.Mol.Biol., 358, 2006
|
|
1YFL
 
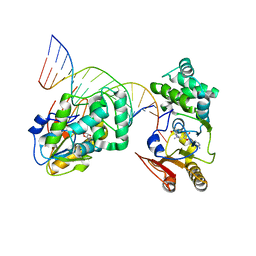 | | T4Dam in Complex with Sinefungin and 16-mer Oligonucleotide Showing Semi-specific and Specific Contact and Flipped Base | | 分子名称: | 5'-D(P*TP*CP*AP*CP*AP*GP*GP*AP*TP*CP*CP*TP*GP*TP*GP*A)-3', DNA adenine methylase, SINEFUNGIN | | 著者 | Horton, J.R, Liebert, K, Hattman, S, Jeltsch, A, Cheng, X. | | 登録日 | 2005-01-03 | | 公開日 | 2005-05-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | Transition from Nonspecific to Specific DNA Interactions along the Substrate-Recognition Pathway of Dam Methyltransferase.
Cell(Cambridge,Mass.), 121, 2005
|
|
1YFJ
 
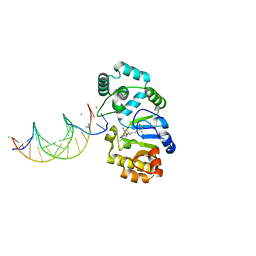 | | T4Dam in Complex with AdoHcy and 15-mer Oligonucleotide Showing Semi-specific and Specific Contact | | 分子名称: | 5'-D(*TP*CP*AP*CP*AP*GP*GP*AP*TP*CP*CP*TP*GP*TP*G)-3', CALCIUM ION, CHLORIDE ION, ... | | 著者 | Horton, J.R, Liebert, K, Hattman, S, Jeltsch, A, Cheng, X. | | 登録日 | 2005-01-02 | | 公開日 | 2005-05-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Transition from Nonspecific to Specific DNA Interactions along the Substrate-Recognition Pathway of Dam Methyltransferase.
Cell(Cambridge,Mass.), 121, 2005
|
|
1YF3
 
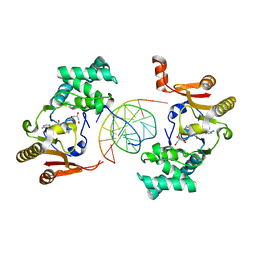 | | T4Dam in Complex with AdoHcy and 13-mer Oligonucleotide Making Non- and Semi-specific (~1/4) Contact | | 分子名称: | 5'-D(*AP*CP*CP*AP*TP*GP*AP*TP*CP*TP*GP*AP*C)-3', 5'-D(*TP*GP*TP*CP*AP*GP*AP*TP*CP*AP*TP*GP*G)-3', DNA adenine methylase, ... | | 著者 | Horton, J.R, Liebert, K, Hattman, S, Jeltsch, A, Cheng, X. | | 登録日 | 2004-12-30 | | 公開日 | 2005-05-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Transition from Nonspecific to Specific DNA Interactions along the Substrate-Recognition Pathway of Dam Methyltransferase.
Cell(Cambridge,Mass.), 121, 2005
|
|
6VDB
 
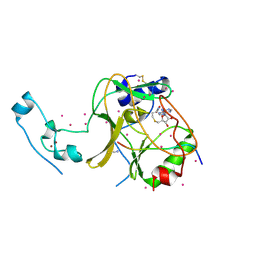 | | SETD2 in complex with a H3-variant super-substrate peptide | | 分子名称: | ALA-PRO-ARG-PHE-GLY-GLY-VAL-MET-ARG-PRO-ASN-ARG, Histone-lysine N-methyltransferase SETD2, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | Beldar, S, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Jeltsch, A, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | 登録日 | 2019-12-24 | | 公開日 | 2020-01-29 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Sequence specificity analysis of the SETD2 protein lysine methyltransferase and discovery of a SETD2 super-substrate.
Commun Biol, 3, 2020
|
|
3EU9
 
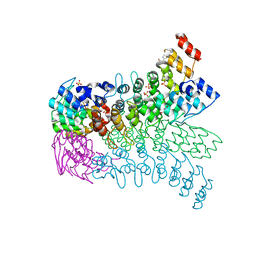 | | The ankyrin repeat domain of Huntingtin interacting protein 14 | | 分子名称: | GLYCEROL, HISTIDINE, Huntingtin-interacting protein 14, ... | | 著者 | Gao, T, Collins, R.E, Horton, J.R, Zhang, R, Zhang, X, Cheng, X. | | 登録日 | 2008-10-09 | | 公開日 | 2009-06-23 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | The ankyrin repeat domain of Huntingtin interacting protein 14 contains a surface aromatic cage, a potential site for methyl-lysine binding.
Proteins, 76, 2009
|
|
1K0Z
 
 | | Crystal Structure of the PvuII endonuclease with Pr3+ and SO4 ions bound in the active site at 2.05A. | | 分子名称: | PRASEODYMIUM ION, SULFATE ION, Type II restriction enzyme PvuII | | 著者 | Spyridaki, A, Athanasiadis, A, Matzen, C, Lanio, T, Jeltsch, A, Simoncsits, A, Scheuring-Vanamee, E, Kokkinidis, M, Pingoud, A. | | 登録日 | 2001-09-21 | | 公開日 | 2003-06-17 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 |
|
|
6W8W
 
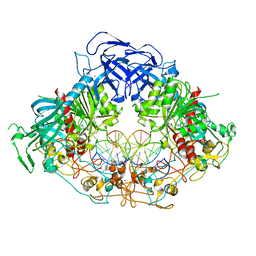 | | Crystal structure of mouse DNMT1 in complex with CCG DNA | | 分子名称: | CCG DNA (5'-D(*AP*CP*TP*TP*AP*(5CM)P*GP*GP*AP*AP*GP*G)-3'), CCG DNA (5'-D(*CP*CP*TP*TP*CP*(C49)P*GP*TP*AP*AP*GP*T)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | 著者 | Anteneh, H, Song, J. | | 登録日 | 2020-03-21 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | DNMT1 activity, base flipping mechanism and genome-wide DNA methylation are regulated by the DNA sequence context
Nat Commun, 2020
|
|
5WCJ
 
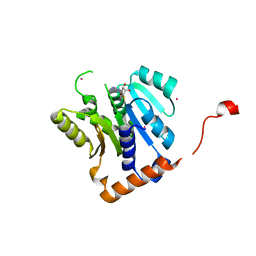 | | Crystal Structure of Human Methyltransferase-like protein 13 in complex with SAH | | 分子名称: | Methyltransferase-like protein 13, S-ADENOSYL-L-HOMOCYSTEINE, UNKNOWN ATOM OR ION | | 著者 | Halabelian, L, Loppnau, P, Seitova, A, Hutchinson, A, Hunt, B, Dong, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | 登録日 | 2017-06-30 | | 公開日 | 2017-07-19 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The dual methyltransferase METTL13 targets N terminus and Lys55 of eEF1A and modulates codon-specific translation rates.
Nat Commun, 9, 2018
|
|
2ORE
 
 | |
6WZZ
 
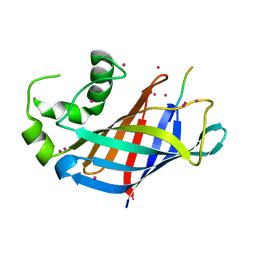 | | GID4 in complex with VGLWKS peptide | | 分子名称: | Glucose-induced degradation protein 4 homolog, UNKNOWN ATOM OR ION, VGLWKS peptide | | 著者 | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2020-05-14 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Recognition of nonproline N-terminal residues by the Pro/N-degron pathway.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WZX
 
 | | GID4 in complex with IGLWKS peptide | | 分子名称: | Glucose-induced degradation protein 4 homolog, ILE-GLY-LEU-TRP-LYS peptide, UNKNOWN ATOM OR ION | | 著者 | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2020-05-14 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Recognition of nonproline N-terminal residues by the Pro/N-degron pathway.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7XYT
 
 | |
2QRV
 
 | | Structure of Dnmt3a-Dnmt3L C-terminal domain complex | | 分子名称: | DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Jia, D, Cheng, X. | | 登録日 | 2007-07-29 | | 公開日 | 2007-12-04 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation.
Nature, 449, 2007
|
|
6BHG
 
 | | Crystal structure of SETDB1 with a modified H3 peptide | | 分子名称: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, UNKNOWN ATOM OR ION | | 著者 | Qin, S, Tempel, W, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2017-10-30 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
5LV4
 
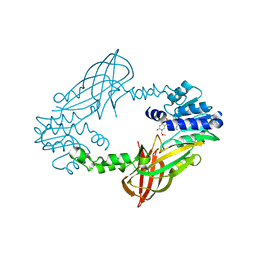 | | Crystal structure of mouse PRMT6 in complex with inhibitor LH1236 | | 分子名称: | (2~{R})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[[4-azanyl-1-(methoxymethyl)-2-oxidanylidene-pyrimidin-5-yl]methyl]amino]-2-azanyl-butanoic acid, Protein arginine N-methyltransferase 6 | | 著者 | Cura, V, Marechal, N, Troffer-Charlier, N, Halby, L, Arimondo, P, Bonnefond, L, Cavarelli, J. | | 登録日 | 2016-09-12 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 373, 2018
|
|
5LV3
 
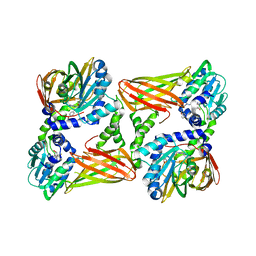 | | Crystal structure of mouse CARM1 in complex with ligand LH1561Br | | 分子名称: | 5-[[2-[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]ethylamino]methyl]-4-azanyl-1-[2-(4-bromanylphenoxy)ethyl]pyrimidin-2-one, Histone-arginine methyltransferase CARM1, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Cura, V, Marechal, N, Troffer-Charlier, N, Halby, L, Arimondo, P, Bonnefond, L, Cavarelli, J. | | 登録日 | 2016-09-12 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 373, 2018
|
|
5LV2
 
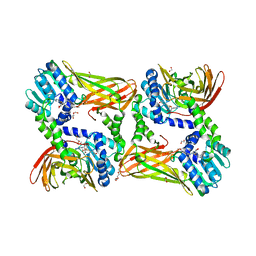 | | Crystal structure of mouse CARM1 in complex with inhibitor LH1246 | | 分子名称: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, ... | | 著者 | Cura, V, Marechal, N, Troffer-Charlier, N, Halby, L, Arimondo, P, Bonnefond, L, Cavarelli, J. | | 登録日 | 2016-09-12 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors.
Philos.Trans.R.Soc.Lond.B Biol.Sci., 373, 2018
|
|
5LV5
 
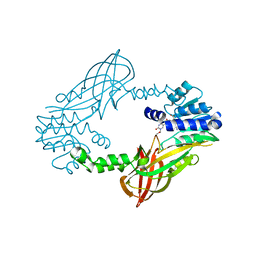 | | Crystal structure of mouse PRMT6 in complex with inhibitor LH1458 | | 分子名称: | 2-[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]ethyl-[[4-azanyl-1-(methoxymethyl)-2-oxidanylidene-pyrimidin-5-yl]methyl]-[(3~{S})-3-azanyl-4-oxidanyl-4-oxidanylidene-butyl]azanium, Protein arginine N-methyltransferase 6 | | 著者 | Cura, V, Marechal, N, Troffer-Charlier, N, Halby, L, Arimondo, P, Bonnefond, L, Cavarelli, J. | | 登録日 | 2016-09-12 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.802 Å) | | 主引用文献 | Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 373, 2018
|
|
5TBJ
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor LH1452 | | 分子名称: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ... | | 著者 | Cura, V, Marechal, N, Troffer-Charlier, N, Halby, L, Arimondo, P, Bonnefond, L, Cavarelli, J. | | 登録日 | 2016-09-12 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors.
Philos.Trans.R.Soc.Lond.B Biol.Sci., 373, 2018
|
|
5TBI
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor LH1427 | | 分子名称: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 4-[2-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylamino]ethylamino]-1-(methoxymethyl)pyrimidin-2-one, ... | | 著者 | Cura, V, Marechal, N, Troffer-Charlier, N, Halby, L, Arimondo, P, Bonnefond, L, Cavarelli, J. | | 登録日 | 2016-09-12 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.294 Å) | | 主引用文献 | Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors.
Philos.Trans.R.Soc.Lond.B Biol.Sci., 373, 2018
|
|
6BHI
 
 | | Crystal structure of SETDB1 with a modified H3 peptide | | 分子名称: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, UNKNOWN ATOM OR ION | | 著者 | Qin, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2017-10-30 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
6BHD
 
 | | Crystal structure of SETDB1 with a modified H3 peptide | | 分子名称: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, SODIUM ION, ... | | 著者 | Qin, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2017-10-30 | | 公開日 | 2017-12-06 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
