2QLD
 
 | | human Hsp40 Hdj1 | | 分子名称: | DnaJ homolog subfamily B member 1 | | 著者 | Hu, J, Wu, Y, Li, J, Fu, Z, Sha, B. | | 登録日 | 2007-07-12 | | 公開日 | 2008-07-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The crystal structure of the putative peptide-binding fragment from the human Hsp40 protein Hdj1.
Bmc Struct.Biol., 8, 2008
|
|
6DWB
 
 | | Structure of the Salmonella SPI-1 type III secretion injectisome needle filament | | 分子名称: | Protein PrgI | | 著者 | Hu, J, Hong, C, Worrall, L.J, Vuckovic, M, Yu, Z, Strynadka, N.C.J. | | 登録日 | 2018-06-26 | | 公開日 | 2018-10-03 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM analysis of the T3S injectisome reveals the structure of the needle and open secretin.
Nat Commun, 9, 2018
|
|
6DV3
 
 | | Structure of the Salmonella SPI-1 type III secretion injectisome secretin InvG in the open gate state | | 分子名称: | Protein InvG | | 著者 | Hu, J, Worrall, L.J, Vuckovic, M, Atkinson, C.E, Strynadka, N.C.J. | | 登録日 | 2018-06-22 | | 公開日 | 2018-10-03 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Cryo-EM analysis of the T3S injectisome reveals the structure of the needle and open secretin.
Nat Commun, 9, 2018
|
|
6DV6
 
 | | Structure of the Salmonella SPI-1 type III secretion injectisome secretin InvG (residues 176-end) in the open gate state | | 分子名称: | Protein InvG | | 著者 | Hu, J, Worrall, L.J, Vuckovic, M, Atkinson, C.E, Strynadka, N.C.J. | | 登録日 | 2018-06-22 | | 公開日 | 2018-10-03 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-EM analysis of the T3S injectisome reveals the structure of the needle and open secretin.
Nat Commun, 9, 2018
|
|
6DUZ
 
 | | Structure of the periplasmic domains of PrgH and PrgK from the assembled Salmonella type III secretion injectisome needle complex | | 分子名称: | Lipoprotein PrgK, Protein PrgH | | 著者 | Hu, J, Worrall, L.J, Vuckovic, M, Atkinson, C.E, Strynadka, N.C.J. | | 登録日 | 2018-06-22 | | 公開日 | 2018-10-03 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM analysis of the T3S injectisome reveals the structure of the needle and open secretin.
Nat Commun, 9, 2018
|
|
1RPY
 
 | | CRYSTAL STRUCTURE OF THE DIMERIC SH2 DOMAIN OF APS | | 分子名称: | SULFATE ION, adaptor protein APS | | 著者 | Hu, J, Liu, J, Ghirlando, R, Saltiel, A.R, Hubbard, S.R. | | 登録日 | 2003-12-03 | | 公開日 | 2003-12-23 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for recruitment of the adaptor protein APS to the activated insulin receptor.
Mol.Cell, 12, 2003
|
|
1RQQ
 
 | | Crystal Structure of the Insulin Receptor Kinase in Complex with the SH2 Domain of APS | | 分子名称: | BISUBSTRATE INHIBITOR, Insulin receptor, MANGANESE (II) ION, ... | | 著者 | Hu, J, Liu, J, Ghirlando, R, Saltiel, A.R, Hubbard, S.R. | | 登録日 | 2003-12-06 | | 公開日 | 2003-12-30 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for recruitment of the adaptor protein APS to the activated insulin receptor.
Mol.Cell, 12, 2003
|
|
6PEP
 
 | | Focussed refinement of InvGN0N1:SpaPQR:PrgIJ from the Salmonella SPI-1 injectisome needle complex | | 分子名称: | Protein InvG, Protein PrgH, Protein PrgI, ... | | 著者 | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | 登録日 | 2019-06-20 | | 公開日 | 2019-10-23 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
6PEE
 
 | |
6PEM
 
 | | Focussed refinement of InvGN0N1:SpaPQR:PrgHK from Salmonella SPI-1 injectisome NC-base | | 分子名称: | Lipoprotein PrgK, Protein InvG, Protein PrgH, ... | | 著者 | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | 登録日 | 2019-06-20 | | 公開日 | 2019-10-23 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
6Q15
 
 | | Structure of the Salmonella SPI-1 injectisome needle complex | | 分子名称: | Lipoprotein PrgK, Protein InvG, Protein PrgH, ... | | 著者 | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | 登録日 | 2019-08-02 | | 公開日 | 2019-10-23 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (5.15 Å) | | 主引用文献 | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
6Q14
 
 | | Structure of the Salmonella SPI-1 injectisome NC-base | | 分子名称: | Lipoprotein PrgK, Protein InvG, Protein PrgH, ... | | 著者 | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | 登録日 | 2019-08-02 | | 公開日 | 2019-10-23 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
6Q16
 
 | | Focussed refinement of InvGN0N1:PrgHK:SpaPQR:PrgIJ from Salmonella SPI-1 injectisome NC-base | | 分子名称: | Lipoprotein PrgK, Protein InvG, Protein PrgH, ... | | 著者 | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | 登録日 | 2019-08-02 | | 公開日 | 2019-10-23 | | 最終更新日 | 2020-01-15 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
3IO3
 
 | | GEt3 with ADP from D. Hansenii in Closed form | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DEHA2D07832p, GLYCEROL, ... | | 著者 | Hu, J, Li, J, Qian, X, Sha, B. | | 登録日 | 2009-08-13 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The crystal structures of yeast Get3 suggest a mechanism for tail-anchored protein membrane insertion
Plos One, 4, 2009
|
|
1YVH
 
 | |
3H84
 
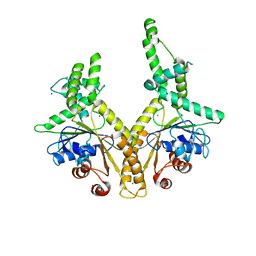 | | Crystal structure of GET3 | | 分子名称: | ATPase GET3, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Hu, J, Li, J, Qian, X, Sha, B. | | 登録日 | 2009-04-28 | | 公開日 | 2009-12-22 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The crystal structures of yeast Get3 suggest a mechanism for tail-anchored protein membrane insertion.
Plos One, 4, 2009
|
|
4TZ7
 
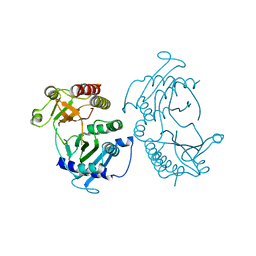 | | Crystal structure of type I phosphatidylinositol 4-phosphate 5-kinase alpha from Zebrafish | | 分子名称: | Phosphatidylinositol-4-phosphate 5-kinase, type I, alpha | | 著者 | Hu, J, Qin, Y, Wang, J, Li, L, Wu, D, Ha, Y. | | 登録日 | 2014-07-09 | | 公開日 | 2015-09-02 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.31 Å) | | 主引用文献 | Resolution of structure of PIP5K1A reveals molecular mechanism for its regulation by dimerization and dishevelled.
Nat Commun, 6, 2015
|
|
9B8Y
 
 | |
9B93
 
 | |
9B92
 
 | |
9B91
 
 | |
9B8W
 
 | |
9B90
 
 | |
9B8Z
 
 | |
9B8X
 
 | |
