5VY8
 
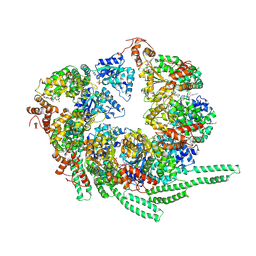 | | S. cerevisiae Hsp104-ADP complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Heat shock protein 104 | | 著者 | Gates, S.N, Yokom, A.L, Lin, J.-B, Jackrel, M.E, Rizo, A.N, Kendsersky, N.M, Buell, C.E, Sweeny, E.A, Chuang, E, Torrente, M.P, Mack, K.L, Su, M, Shorter, J, Southworth, D.R. | | 登録日 | 2017-05-24 | | 公開日 | 2017-07-05 | | 最終更新日 | 2018-08-15 | | 実験手法 | ELECTRON MICROSCOPY (5.6 Å) | | 主引用文献 | Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104.
Science, 357, 2017
|
|
5VYA
 
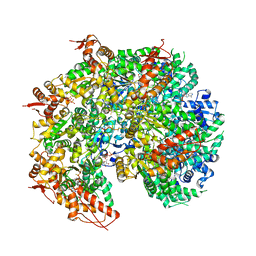 | | S. cerevisiae Hsp104:casein complex, Extended Conformation | | 分子名称: | Alpha-S1-casein, Heat shock protein 104, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | 著者 | Gates, S.N, Yokom, A.L, Lin, J.-B, Jackrel, M.E, Rizo, A.N, Kendsersky, N.M, Buell, C.E, Sweeny, E.A, Chuang, E, Torrente, M.P, Mack, K.L, Su, M, Shorter, J, Southworth, D.R. | | 登録日 | 2017-05-24 | | 公開日 | 2017-07-05 | | 最終更新日 | 2017-08-02 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104.
Science, 357, 2017
|
|
5VY9
 
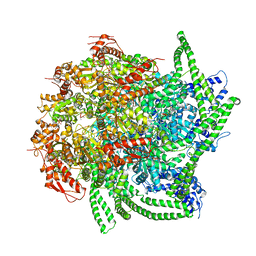 | | S. cerevisiae Hsp104:casein complex, Middle Domain Conformation | | 分子名称: | Alpha-S1-casein, Heat shock protein 104, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | 著者 | Gates, S.N, Yokom, A.L, Lin, J.-B, Jackrel, M.E, Rizo, A.N, Kendsersky, N.M, Buell, C.E, Sweeny, E.A, Chuang, E, Torrente, M.P, Mack, K.L, Su, M, Shorter, J, Southworth, D.R. | | 登録日 | 2017-05-24 | | 公開日 | 2017-07-19 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (6.7 Å) | | 主引用文献 | Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104.
Science, 357, 2017
|
|
5VJH
 
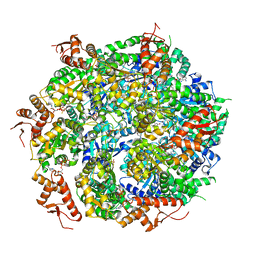 | | Closed State CryoEM Reconstruction of Hsp104:ATPyS and FITC casein | | 分子名称: | FITC casein, Heat shock protein 104, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | 著者 | Gates, S.N, Yokom, A.L, Lin, J.-B, Jackrel, M.E, Rizo, A.N, Kendsersky, N.M, Buell, C.E, Sweeny, E.A, Chuang, E, Torrente, M.P, Mack, K.L, Su, M, Shorter, J, Southworth, D.R. | | 登録日 | 2017-04-19 | | 公開日 | 2017-07-05 | | 最終更新日 | 2017-08-02 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104.
Science, 357, 2017
|
|
5KNE
 
 | | CryoEM Reconstruction of Hsp104 Hexamer | | 分子名称: | Heat shock protein 104, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Yokom, A.L, Gates, S.N, Jackrel, M.E, Mack, K.L, Su, M, Shorter, J, Southworth, D.R. | | 登録日 | 2016-06-28 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (5.64 Å) | | 主引用文献 | Spiral architecture of the Hsp104 disaggregase reveals the basis for polypeptide translocation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
6EF0
 
 | | Yeast 26S proteasome bound to ubiquitinated substrate (1D* motor state) | | 分子名称: | 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6B homolog, ... | | 著者 | de la Pena, A.H, Goodall, E.A, Gates, S.N, Lander, G.C, Martin, A. | | 登録日 | 2018-08-15 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.43 Å) | | 主引用文献 | Substrate-engaged 26Sproteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation.
Science, 362, 2018
|
|
6XF9
 
 | | Crystal structure of KSHV ORF68 | | 分子名称: | Packaging protein UL32, ZINC ION | | 著者 | Didychuk, A.L, Gates, S.N, Martin, A, Glaunsinger, B. | | 登録日 | 2020-06-15 | | 公開日 | 2021-02-24 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | A pentameric protein ring with novel architecture is required for herpesviral packaging.
Elife, 10, 2021
|
|
6XFA
 
 | | Cryo-EM structure of EBV BFLF1 | | 分子名称: | Packaging protein UL32, ZINC ION | | 著者 | Didychuk, A.L, Gates, S.N, Martin, A, Glaunsinger, B. | | 登録日 | 2020-06-15 | | 公開日 | 2021-02-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | A pentameric protein ring with novel architecture is required for herpesviral packaging.
Elife, 10, 2021
|
|
6EF2
 
 | | Yeast 26S proteasome bound to ubiquitinated substrate (5T motor state) | | 分子名称: | 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6B homolog, ... | | 著者 | de la Pena, A.H, Goodall, E.A, Gates, S.N, Lander, G.C, Martin, A. | | 登録日 | 2018-08-15 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.27 Å) | | 主引用文献 | Substrate-engaged 26Sproteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation.
Science, 362, 2018
|
|
6EF3
 
 | | Yeast 26S proteasome bound to ubiquitinated substrate (4D motor state) | | 分子名称: | 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6B homolog, ... | | 著者 | de la Pena, A.H, Goodall, E.A, Gates, S.N, Lander, G.C, Martin, A. | | 登録日 | 2018-08-15 | | 公開日 | 2018-10-17 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (4.17 Å) | | 主引用文献 | Substrate-engaged 26Sproteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation.
Science, 362, 2018
|
|
6EF1
 
 | | Yeast 26S proteasome bound to ubiquitinated substrate (5D motor state) | | 分子名称: | 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6B homolog, ... | | 著者 | de la Pena, A.H, Goodall, E.A, Gates, S.N, Lander, G.C, Martin, A. | | 登録日 | 2018-08-15 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.73 Å) | | 主引用文献 | Substrate-engaged 26Sproteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation.
Science, 362, 2018
|
|
6OG2
 
 | | Focus classification structure of the hyperactive ClpB mutant K476C, bound to casein, post-state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Hyperactive disaggregase ClpB | | 著者 | Rizo, A.R, Lin, J.-B, Gates, S.N, Tse, E, Bart, S.M, Castellano, L.M, Dimaio, F, Shorter, J, Southworth, D.R. | | 登録日 | 2019-04-01 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural basis for substrate gripping and translocation by the ClpB AAA+ disaggregase.
Nat Commun, 10, 2019
|
|
6OG3
 
 | | Focus classification structure of the hyperactive ClpB mutant K476C, bound to casein, NTD-trimer | | 分子名称: | Alpha S1-casein, Hyperactive disaggregase ClpB | | 著者 | Rizo, A.R, Lin, J.-B, Gates, S.N, Tse, E, Bart, S.M, Castellano, L.M, Dimaio, F, Shorter, J, Southworth, D.R. | | 登録日 | 2019-04-01 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural basis for substrate gripping and translocation by the ClpB AAA+ disaggregase.
Nat Commun, 10, 2019
|
|
6OG1
 
 | | Focus classification structure of the hyperactive ClpB mutant K476C, bound to casein, pre-state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Hyperactive disaggregase ClpB, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | 著者 | Rizo, A.R, Lin, J.-B, Gates, S.N, Tse, E, Bart, S.M, Castellano, L.M, Dimaio, F, Shorter, J, Southworth, D.R. | | 登録日 | 2019-04-01 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis for substrate gripping and translocation by the ClpB AAA+ disaggregase.
Nat Commun, 10, 2019
|
|
6OAX
 
 | | Structure of the hyperactive ClpB mutant K476C, bound to casein, pre-state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Alpha-S1-casein, Hyperactive disaggregase ClpB, ... | | 著者 | Rizo, A.R, Lin, J.-B, Gates, S.N, Tse, E, Bart, S.M, Castellano, L.M, Dimaio, F, Shorter, J, Southworth, D.R. | | 登録日 | 2019-03-18 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis for substrate gripping and translocation by the ClpB AAA+ disaggregase.
Nat Commun, 10, 2019
|
|
6OAY
 
 | | Structure of the hyperactive ClpB mutant K476C, bound to casein, post-state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Alpha-S1-casein, Hyperactive disaggregase ClpB, ... | | 著者 | Rizo, A.R, Lin, J.-B, Gates, S.N, Tse, E, Bart, S.M, Castellano, L.M, Dimaio, F, Shorter, J, Southworth, D.R. | | 登録日 | 2019-03-18 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis for substrate gripping and translocation by the ClpB AAA+ disaggregase.
Nat Commun, 10, 2019
|
|
7L7J
 
 | | Cryo-EM structure of Hsp90:p23 closed-state complex | | 分子名称: | Heat shock protein HSP 90-alpha, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Prostaglandin E synthase 3 | | 著者 | Lee, K, Thwin, A.C, Tse, E, Gates, S.N, Southworth, D.R. | | 登録日 | 2020-12-28 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | The structure of an Hsp90-immunophilin complex reveals cochaperone recognition of the client maturation state.
Mol.Cell, 81, 2021
|
|
7L7I
 
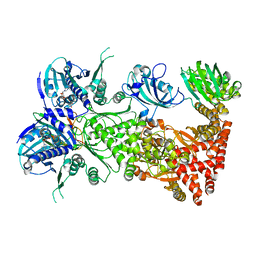 | | Cryo-EM structure of Hsp90:FKBP51:p23 closed-state complex | | 分子名称: | Heat shock protein HSP 90-alpha, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Peptidyl-prolyl cis-trans isomerase FKBP5, ... | | 著者 | Lee, K, Thwin, A.C, Tse, E, Gates, S.N, Southworth, D.R. | | 登録日 | 2020-12-28 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | The structure of an Hsp90-immunophilin complex reveals cochaperone recognition of the client maturation state.
Mol.Cell, 81, 2021
|
|
