3J4J
 
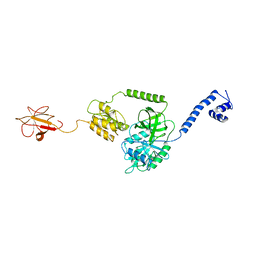 | | Model of full-length T. thermophilus Translation Initiation Factor 2 refined against its cryo-EM density from a 30S Initiation Complex map | | 分子名称: | Translation initiation factor IF-2 | | 著者 | Simonetti, A, Marzi, S, Billas, I.M.L, Tsai, A, Fabbretti, A, Myasnikov, A, Roblin, P, Vaiana, A.C, Hazemann, I, Eiler, D, Steitz, T.A, Puglisi, J.D, Gualerzi, C.O, Klaholz, B.P. | | 登録日 | 2013-08-26 | | 公開日 | 2013-09-25 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (11.5 Å) | | 主引用文献 | Involvement of protein IF2 N domain in ribosomal subunit joining revealed from architecture and function of the full-length initiation factor.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7OI3
 
 | | Cryo-EM structure of the Cetacean morbillivirus nucleoprotein-RNA complex | | 分子名称: | Cetacean morbillivirus nucleoprotein, poly-A 6-mer | | 著者 | Zinzula, L, Beck, F, Klumpe, S, Bohn, S, Pfeifer, G, Bollschweiler, D, Nagy, I, Plitzko, J.M, Baumeister, W. | | 登録日 | 2021-05-11 | | 公開日 | 2021-06-23 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Cryo-EM structure of the cetacean morbillivirus nucleoprotein-RNA complex.
J.Struct.Biol., 213, 2021
|
|
6I3N
 
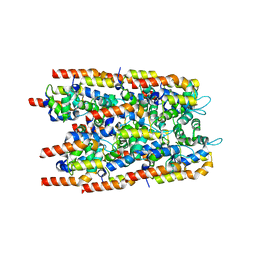 | |
8PKJ
 
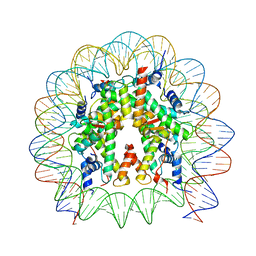 | | Cryo-EM structure of the nucleosome containing Nr5a2 motif at SHL+5.5 | | 分子名称: | DNA, Histone H2A, Histone H2B, ... | | 著者 | Kobayashi, W, Sappler, A, Bollschweiler, D, Kummecke, M, Basquin, J, Arslantas, E, Ruangroengkulrith, S, Hornberger, R, Duderstadt, K, Tachibana, K. | | 登録日 | 2023-06-26 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Nucleosome-bound NR5A2 structure reveals pioneer factor mechanism by DNA minor groove anchor competition.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8PKI
 
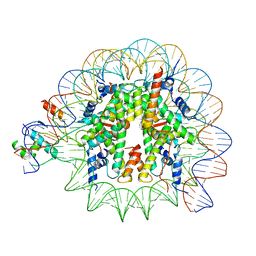 | | Cryo-EM structure of NR5A2-nucleosome complex SHL+5.5 | | 分子名称: | DNA, Histone H2A, Histone H2B type 1-C/E/G, ... | | 著者 | Kobayashi, W, Sappler, A, Bollschweiler, D, Kummecke, M, Basquin, J, Arslantas, E, Ruangroengkulrith, S, Hornberger, R, Duderstadt, K, Tachibana, K. | | 登録日 | 2023-06-26 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.58 Å) | | 主引用文献 | Nucleosome-bound NR5A2 structure reveals pioneer factor mechanism by DNA minor groove anchor competition.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8BSC
 
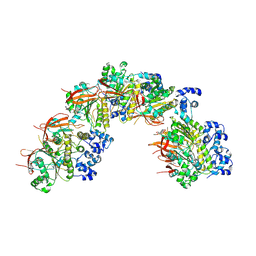 | |
8BQ2
 
 | | CryoEM structure of the pre-synaptic RAD51 nucleoprotein filament in the presence of ATP and Ca2+ | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (30-MER), ... | | 著者 | Appleby, R, Bollschweiler, D, Pellegrini, L. | | 登録日 | 2022-11-18 | | 公開日 | 2023-05-03 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | A metal ion-dependent mechanism of RAD51 nucleoprotein filament disassembly.
Iscience, 26, 2023
|
|
8BR2
 
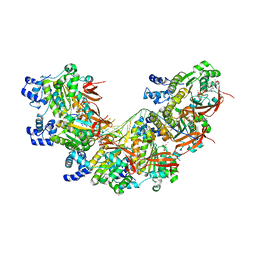 | | CryoEM structure of the post-synaptic RAD51 nucleoprotein filament in the presence of ATP and Ca2+ | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(P*GP*CP*GP*AP*GP*CP*TP*CP*GP*AP*TP*GP*CP*AP*CP*CP*TP*CP*CP*A)-3'), ... | | 著者 | Appleby, R, Bollschweiler, D, Pellegrini, L. | | 登録日 | 2022-11-22 | | 公開日 | 2023-05-03 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | A metal ion-dependent mechanism of RAD51 nucleoprotein filament disassembly.
Iscience, 26, 2023
|
|
6Z6F
 
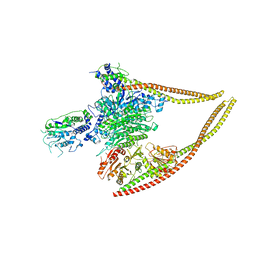 | | HDAC-PC | | 分子名称: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | 著者 | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | 登録日 | 2020-05-28 | | 公開日 | 2021-02-17 | | 実験手法 | ELECTRON MICROSCOPY (3.11 Å) | | 主引用文献 | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
6Z6H
 
 | | HDAC-DC | | 分子名称: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | 著者 | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | 登録日 | 2020-05-28 | | 公開日 | 2021-02-17 | | 実験手法 | ELECTRON MICROSCOPY (8.55 Å) | | 主引用文献 | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
6Z6P
 
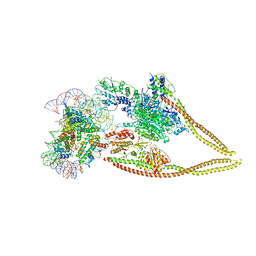 | | HDAC-PC-Nuc | | 分子名称: | DNA (145-MER), HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, ... | | 著者 | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | 登録日 | 2020-05-28 | | 公開日 | 2021-02-17 | | 実験手法 | ELECTRON MICROSCOPY (4.43 Å) | | 主引用文献 | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
6Z6O
 
 | | HDAC-TC | | 分子名称: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | 著者 | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | 登録日 | 2020-05-28 | | 公開日 | 2021-02-17 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
4KJZ
 
 | |
4QJD
 
 | |
4QJH
 
 | |
6DVK
 
 | | Computationally designed mini tetraloop-tetraloop receptor by the RNAMake program - construct 6 (miniTTR 6) | | 分子名称: | COBALT (II) ION, MAGNESIUM ION, RNA (95-MER) | | 著者 | Eiler, D.R, Yesselman, J.D, Costantino, D.A, Das, R, Kieft, J.S. | | 登録日 | 2018-06-24 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Computational design of three-dimensional RNA structure and function.
Nat Nanotechnol, 14, 2019
|
|
4Q6G
 
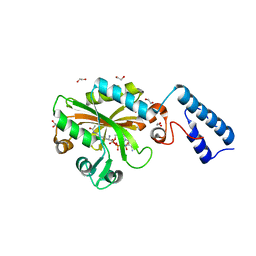 | | Crystal Structure of the C-terminal domain of AcKRS-1 bound with N-acetyl-lysine and ADPNP | | 分子名称: | 1,2-ETHANEDIOL, N(6)-ACETYLLYSINE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Eiler, D.R, Kavran, J, Steitz, T.A. | | 登録日 | 2014-04-22 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Polyspecific pyrrolysyl-tRNA synthetases from directed evolution.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4ZIB
 
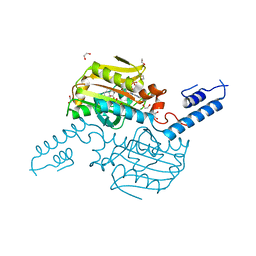 | | Crystal Structure of the C-terminal domain of PylRS mutant bound with 3-benzothienyl-l-alanine and ATP | | 分子名称: | 1,2-ETHANEDIOL, 3-(1-benzothiophen-3-yl)-L-alanine, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Nakamura, A, Guo, L.T, Wang, Y.S, Soll, D. | | 登録日 | 2015-04-28 | | 公開日 | 2016-03-02 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.054 Å) | | 主引用文献 | Probing the active site tryptophan of Staphylococcus aureus thioredoxin with an analog.
Nucleic Acids Res., 43, 2015
|
|
4TQF
 
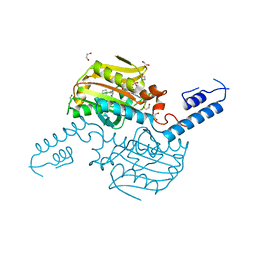 | | Crystal Structure of the C-terminal domain of IFRS bound with 2-(5-bromothienyl)-L-Ala and ATP | | 分子名称: | 1,2-ETHANEDIOL, 3-(5-bromothiophen-2-yl)-L-alanine, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Nakamura, A, O'Donoghue, P, Soll, D. | | 登録日 | 2014-06-11 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.7143 Å) | | 主引用文献 | Polyspecific pyrrolysyl-tRNA synthetases from directed evolution.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TQD
 
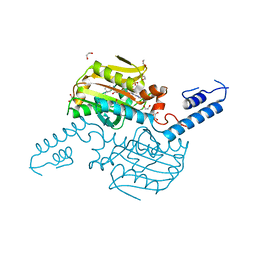 | | Crystal Structure of the C-terminal domain of IFRS bound with 3-iodo-L-Phe and ATP | | 分子名称: | 1,2-ETHANEDIOL, 3-iodo-L-phenylalanine, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Nakamura, A, O'Donoghue, P, Soll, D. | | 登録日 | 2014-06-11 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.1429 Å) | | 主引用文献 | Polyspecific pyrrolysyl-tRNA synthetases from directed evolution.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6SWY
 
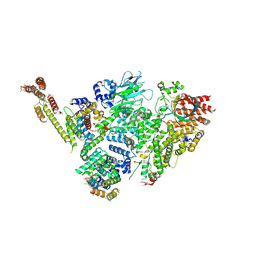 | | Structure of active GID E3 ubiquitin ligase complex minus Gid2 and delta Gid9 RING domain | | 分子名称: | Glucose-induced degradation protein 8, Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10, Vacuolar import and degradation protein 24, ... | | 著者 | Qiao, S, Prabu, J.R, Schulman, B.A. | | 登録日 | 2019-09-24 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Interconversion between Anticipatory and Active GID E3 Ubiquitin Ligase Conformations via Metabolically Driven Substrate Receptor Assembly
Mol.Cell, 77, 2020
|
|
5EQW
 
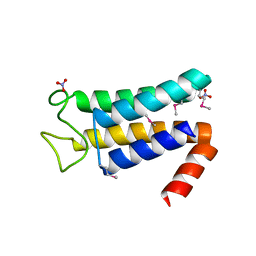 | | Structure of the major structural protein D135 of Acidianus tailed spindle virus (ATSV) | | 分子名称: | NITRATE ION, Putative major coat protein | | 著者 | Hochstein, R.A, Lintner, N.G, Young, M.J, Lawrence, C.M. | | 登録日 | 2015-11-13 | | 公開日 | 2016-11-16 | | 最終更新日 | 2019-11-27 | | 実験手法 | X-RAY DIFFRACTION (1.679 Å) | | 主引用文献 | Structural studies ofAcidianustailed spindle virus reveal a structural paradigm used in the assembly of spindle-shaped viruses.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6YBQ
 
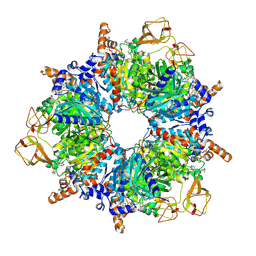 | | Engineered glycolyl-CoA carboxylase (quintuple mutant) with bound CoA | | 分子名称: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, COENZYME A, Propionyl-CoA carboxylase alpha subunit, ... | | 著者 | Schuller, J.M, Schuller, S.K, Zarzycki, J, Scheffen, M, Marchal, D.M, Erb, T.J. | | 登録日 | 2020-03-17 | | 公開日 | 2020-10-28 | | 最終更新日 | 2021-05-12 | | 実験手法 | ELECTRON MICROSCOPY (1.96 Å) | | 主引用文献 | A new-to-nature carboxylation module to improve natural and synthetic CO2 fixation
Nat Catal, 2021
|
|
6YBP
 
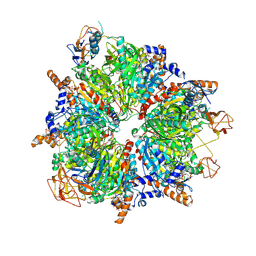 | | Propionyl-CoA carboxylase of Methylorubrum extorquens with bound CoA | | 分子名称: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, COENZYME A, Propionyl-CoA carboxylase alpha subunit, ... | | 著者 | Schuller, J.M, Schuller, S.K, Zarzycki, J, Scheffen, M, Marchal, D.M, Erb, T.J. | | 登録日 | 2020-03-17 | | 公開日 | 2020-10-28 | | 最終更新日 | 2021-05-12 | | 実験手法 | ELECTRON MICROSCOPY (3.48 Å) | | 主引用文献 | A new-to-nature carboxylation module to improve natural and synthetic CO2 fixation
Nat Catal, 2021
|
|
