6MEP
 
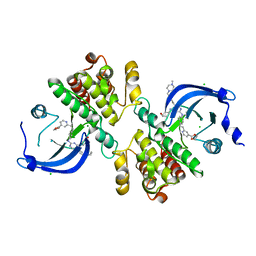 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC3437 | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, Tyrosine-protein kinase Mer, ... | | 著者 | Da, C, Zhang, D, Stashko, M.A, Cheng, A, Hunter, D, Norris-Drouin, J, Graves, L, Machius, M, Miley, M.J, DeRyckere, D, Earp, H.S, Graham, D.K, Frye, S.V, Wang, X, Kireev, D. | | 登録日 | 2018-09-06 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.893 Å) | | 主引用文献 | Data-Driven Construction of Antitumor Agents with Controlled Polypharmacology.
J.Am.Chem.Soc., 141, 2019
|
|
2ETL
 
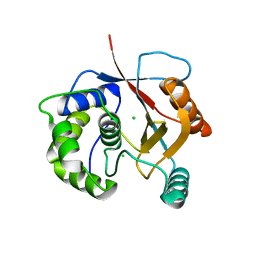 | | Crystal Structure of Ubiquitin Carboxy-terminal Hydrolase L1 (UCH-L1) | | 分子名称: | CHLORIDE ION, Ubiquitin carboxyl-terminal hydrolase isozyme L1 | | 著者 | Das, C, Hoang, Q.Q, Kreinbring, C.A, Luchansky, S.J, Meray, R.K, Ray, S.S, Lansbury, P.T, Ringe, D, Petsko, G.A. | | 登録日 | 2005-10-27 | | 公開日 | 2006-03-28 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for conformational plasticity of the Parkinson's disease-associated ubiquitin hydrolase UCH-L1.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
3RII
 
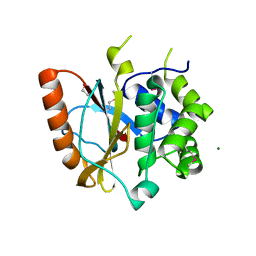 | | Crystal structure of the catalytic domain of UCHL5, a proteasome-associated human deubiquitinating enzyme, reveals an unproductive form of the enzyme | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | 著者 | Das, C. | | 登録日 | 2011-04-13 | | 公開日 | 2011-11-09 | | 最終更新日 | 2011-12-14 | | 実験手法 | X-RAY DIFFRACTION (2.0008 Å) | | 主引用文献 | Crystal structure of the catalytic domain of UCHL5, a proteasome-associated human deubiquitinating enzyme, reveals an unproductive form of the enzyme.
Febs J., 278, 2011
|
|
3RIS
 
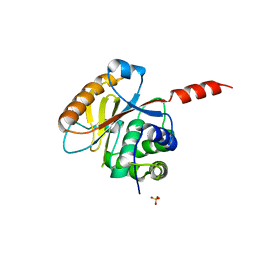 | | Crystal structure of the catalytic domain of UCHL5, a proteasome-associated human deubiquitinating enzyme, reveals an unproductive form of the enzyme | | 分子名称: | GLYCEROL, SULFATE ION, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | 著者 | Das, C, Permaul, M, Maiti, T.K. | | 登録日 | 2011-04-14 | | 公開日 | 2011-11-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.398 Å) | | 主引用文献 | Crystal structure of the catalytic domain of UCHL5, a proteasome-associated human deubiquitinating enzyme, reveals an unproductive form of the enzyme.
Febs J., 278, 2011
|
|
3IFW
 
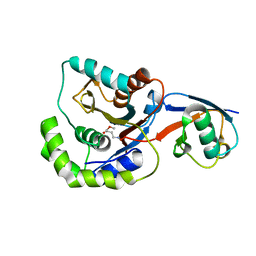 | |
4IG7
 
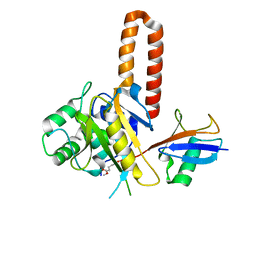 | |
4OUF
 
 | | Crystal Structure of CBP bromodomain | | 分子名称: | 1,2-ETHANEDIOL, CREB-binding protein, DI(HYDROXYETHYL)ETHER | | 著者 | Roy, S, Das, C, Tyler, J.K, Kutateladze, T.G. | | 登録日 | 2014-02-17 | | 公開日 | 2014-03-12 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Binding of the histone chaperone ASF1 to the CBP bromodomain promotes histone acetylation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4PQT
 
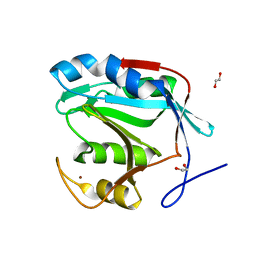 | | Insights into the mechanism of deubiquitination by JAMM deubiquitinases from co-crystal structures of enzyme with substrate and product | | 分子名称: | 1,2-ETHANEDIOL, AMSH-like protease sst2, Protein UBBP4, ... | | 著者 | Shrestha, R.K, Ronau, J.A, Das, C. | | 登録日 | 2014-03-04 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Insights into the Mechanism of Deubiquitination by JAMM Deubiquitinases from Cocrystal Structures of the Enzyme with the Substrate and Product.
Biochemistry, 53, 2014
|
|
7LM3
 
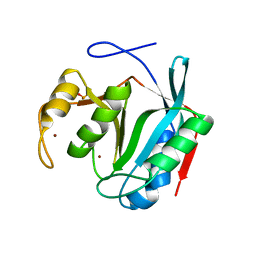 | |
4Q3Y
 
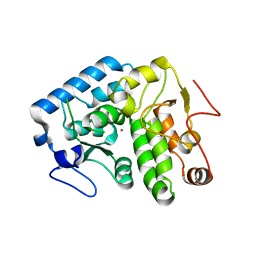 | |
4Q3Z
 
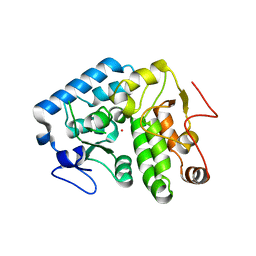 | |
4Q3W
 
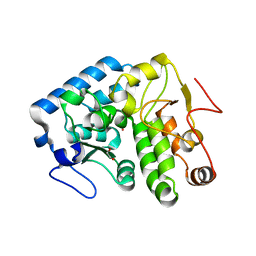 | | Crystal structure of C. violaceum phenylalanine hydroxylase D139E mutation | | 分子名称: | 1,2-ETHANEDIOL, COBALT (II) ION, Phenylalanine-4-hydroxylase | | 著者 | Ronau, J.A, Abu-Omar, M.M, Das, C. | | 登録日 | 2014-04-12 | | 公開日 | 2015-02-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | A conserved acidic residue in phenylalanine hydroxylase contributes to cofactor affinity and catalysis.
Biochemistry, 53, 2014
|
|
4Q3X
 
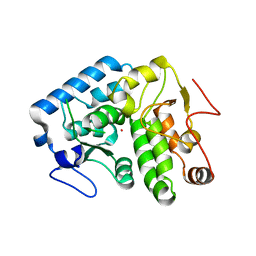 | |
6WTG
 
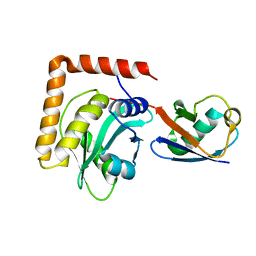 | | SdeA DUB Domain in complex with Ubiquitin | | 分子名称: | Ubiquitin, Ubiquitinating/deubiquitinating enzyme SdeA | | 著者 | Kenny, S, Sheedlo, M, Das, C. | | 登録日 | 2020-05-02 | | 公開日 | 2021-03-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | Insights into Ubiquitin Product Release in Hydrolysis Catalyzed by the Bacterial Deubiquitinase SdeA.
Biochemistry, 60, 2021
|
|
8FEK
 
 | |
6MRN
 
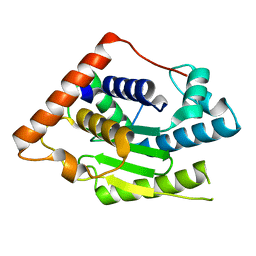 | | Crystal Structure of ChlaDUB2 DUB domain | | 分子名称: | Deubiquitinase and deneddylase Dub2 | | 著者 | Hausman, J.M, Das, C. | | 登録日 | 2018-10-15 | | 公開日 | 2019-10-30 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | The Two Deubiquitinating Enzymes fromChlamydia trachomatisHave Distinct Ubiquitin Recognition Properties.
Biochemistry, 59, 2020
|
|
8UX2
 
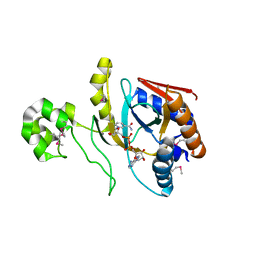 | | Chromobacterium violaceum mono-ADP-ribosyltransferase CteC in complex with NAD+ | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, NAD(+)--protein-threonine ADP-ribosyltransferase, ... | | 著者 | Zhang, Z, Rondon, H, Das, C. | | 登録日 | 2023-11-08 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Crystal structure of bacterial ubiquitin ADP-ribosyltransferase CteC reveals a substrate-recruiting insertion.
J.Biol.Chem., 300, 2023
|
|
8DMU
 
 | |
5KKV
 
 | |
8DMR
 
 | |
6OAM
 
 | | Crystal Structure of ChlaDUB2 DUB domain | | 分子名称: | Deubiquitinase and deneddylase Dub2, Ubiquitin | | 著者 | Hausman, J.M, Das, C. | | 登録日 | 2019-03-17 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.503 Å) | | 主引用文献 | The Two Deubiquitinating Enzymes fromChlamydia trachomatisHave Distinct Ubiquitin Recognition Properties.
Biochemistry, 59, 2020
|
|
6OV1
 
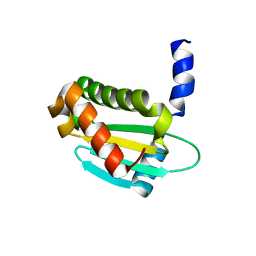 | | Structure of Staphylococcus aureus RNase P protein mutant with defective mRNA degradation activity | | 分子名称: | Ribonuclease P protein component | | 著者 | Ha, L, Colquhoun, J, Noinaj, N, Das, C, Dunman, P, Flaherty, D.P. | | 登録日 | 2019-05-06 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Genetic and biochemical characterization of Staphylococcus aureus RnpA
To Be Published
|
|
6P5H
 
 | |
6P5B
 
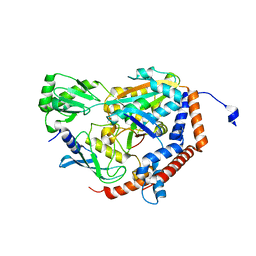 | | Crystal Structure of MavC in Complex with Ub-UbE2N | | 分子名称: | MavC, Ubiquitin, Ubiquitin-conjugating enzyme E2 N | | 著者 | Puvar, K, Iyer, S, Negron Teron, K.I, Das, C. | | 登録日 | 2019-05-30 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.099 Å) | | 主引用文献 | Legionella effector MavC targets the Ube2N~Ub conjugate for noncanonical ubiquitination.
Nat Commun, 11, 2020
|
|
8DQF
 
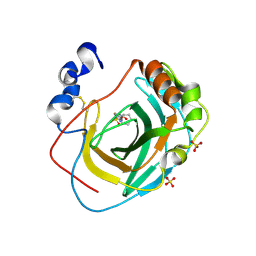 | | Crystal structure of Neisseria gonorrhoeae carbonic anhydrase with N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)cyclohexanecarboxamide | | 分子名称: | Carbonic anhydrase, N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)cyclohexanecarboxamide, SULFATE ION, ... | | 著者 | Marapaka, A.K, Das, C, Flaherty, D.P, Yadav, R. | | 登録日 | 2022-07-19 | | 公開日 | 2022-12-14 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural Characterization of Thiadiazolesulfonamide Inhibitors Bound to Neisseria gonorrhoeae alpha-Carbonic Anhydrase.
Acs Med.Chem.Lett., 14, 2023
|
|
