7OYH
 
 | |
7OY3
 
 | |
7OXY
 
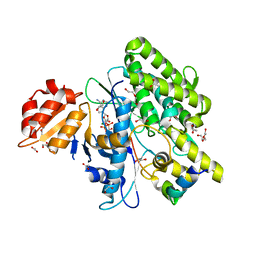 | | Crystal structure of depupylase Dop in complex with Pup and AMP-PCP | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Cui, H. | | 登録日 | 2021-06-23 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structures of prokaryotic ubiquitin-like protein Pup in complex with depupylase Dop reveal the mechanism of catalytic phosphate formation.
Nat Commun, 12, 2021
|
|
7OXV
 
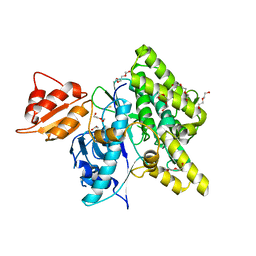 | | Crystal structure of depupylase Dop in the Dop-loop-inserted state | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Depupylase, ... | | 著者 | Cui, H. | | 登録日 | 2021-06-23 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.394 Å) | | 主引用文献 | Structures of prokaryotic ubiquitin-like protein Pup in complex with depupylase Dop reveal the mechanism of catalytic phosphate formation.
Nat Commun, 12, 2021
|
|
7OYF
 
 | |
7R9C
 
 | | Cocrystal of BRD4(D1) with N,N-dimethyl-2-[(3R)-3-(5-{2-[2-methyl-5-(propan-2-yl)phenoxy]pyrimidin-4-yl}-4-[4-(trifluoromethyl)phenyl]-1H-imidazol-1-yl)pyrrolidin-1-yl]ethan-1-amine | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, CHLORIDE ION, ... | | 著者 | Cui, H, Shi, K, Aihara, H, Pomerantz, W.C.K. | | 登録日 | 2021-06-29 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
7RXS
 
 | | Crystal of BRD4(D1) with 2-[(3S)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine | | 分子名称: | 1,2-ETHANEDIOL, 2-[(3S)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine, Bromodomain-containing protein 4 | | 著者 | Cui, H, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | 登録日 | 2021-08-23 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
7RXT
 
 | | Crystal of BRD4(D1) with 2-[(3R)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine | | 分子名称: | 2-[(3R)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine, Bromodomain-containing protein 4 | | 著者 | Cui, H, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | 登録日 | 2021-08-23 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
7RXR
 
 | | Crystal Structure of BRD4(D1) with 4-[4-(4-bromophenyl)-1-(piperidin-4-yl)-1H-imidazol-5-yl]-N-(3,5-dimethylphenyl)pyrimidin-2-amine | | 分子名称: | 1,2-ETHANEDIOL, 4-[4-(4-bromophenyl)-1-(piperidin-4-yl)-1H-imidazol-5-yl]-N-(3,5-dimethylphenyl)pyrimidin-2-amine, Bromodomain-containing protein 4 | | 著者 | Cui, H, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | 登録日 | 2021-08-23 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
6TZW
 
 | |
7MLS
 
 | | X-ray crystal structure of human BRD4(D1) in complex with 2-(2,5-dibromophenoxy)-6-[4-methyl-1-(piperidin-4-yl)-1H-1,2,3-triazol-5-yl]pyridine (compound 23) | | 分子名称: | 1,2-ETHANEDIOL, 2-(2,5-dibromophenoxy)-6-[4-methyl-1-(piperidin-4-yl)-1H-1,2,3-triazol-5-yl]pyridine, Bromodomain-containing protein 4, ... | | 著者 | Cui, H, Johnson, J.A, Zahid, H, Buchholz, C.R, Shi, K, Aihara, H, Pomerantz, W.C.K. | | 登録日 | 2021-04-28 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.26 Å) | | 主引用文献 | 4-Methyl-1,2,3-Triazoles as N -Acetyl-Lysine Mimics Afford Potent BET Bromodomain Inhibitors with Improved Selectivity.
J.Med.Chem., 64, 2021
|
|
7MLR
 
 | | X-ray crystal structure of human BRD4(D1) in complex with 2-(4-{5-[6-(3,5-dimethylphenoxy)pyridin-2-yl]-4-methyl-1H-1,2,3-triazol-1- yl}piperidin-1-yl)-N,N-dimethylethan-1-amine (DW34) | | 分子名称: | 1,2-ETHANEDIOL, 2-(4-{5-[6-(3,5-dimethylphenoxy)pyridin-2-yl]-4-methyl-1H-1,2,3-triazol-1-yl}piperidin-1-yl)-N,N-dimethylethan-1-amine, Bromodomain-containing protein 4, ... | | 著者 | Cui, H, Shi, K, Aihara, H, Pomerantz, W.C.K. | | 登録日 | 2021-04-28 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | 4-Methyl-1,2,3-Triazoles as N -Acetyl-Lysine Mimics Afford Potent BET Bromodomain Inhibitors with Improved Selectivity.
J.Med.Chem., 64, 2021
|
|
7MLQ
 
 | | X-ray crystal structure of human BRD4(D1) in complex with 2-(4-{5-[6-(2,5-dibromophenoxy)pyridin-2-yl]-4-methyl-1H-1,2,3-triazol-1-yl}piperidin-1-yl)-N,N-dimethylethan-1-amine (compound 26) | | 分子名称: | 1,2-ETHANEDIOL, 2-(4-{5-[6-(2,5-dibromophenoxy)pyridin-2-yl]-4-methyl-1H-1,2,3-triazol-1-yl}piperidin-1-yl)-N,N-dimethylethan-1-amine, Bromodomain-containing protein 4, ... | | 著者 | Cui, H, Johnson, J.A, Vail, N.R, Shi, K, Aihara, H, Pomerantz, W.C.K. | | 登録日 | 2021-04-28 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | 4-Methyl-1,2,3-Triazoles as N -Acetyl-Lysine Mimics Afford Potent BET Bromodomain Inhibitors with Improved Selectivity.
J.Med.Chem., 64, 2021
|
|
6WGX
 
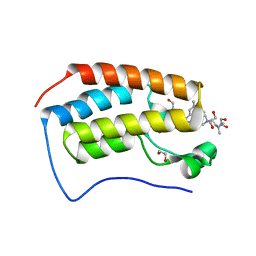 | | Cocrystal of BRD4(D1) with a selective inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 4-(1-{1-[2-(dimethylamino)ethyl]piperidin-4-yl}-4-[4-(trifluoromethyl)phenyl]-1H-imidazol-5-yl)-N-(3,5-dimethylphenyl)pyrimidin-2-amine, Bromodomain-containing protein 4 | | 著者 | Johnson, J.A, Cui, H, Shi, K, Aihara, H, Pomerantz, W.C.K. | | 登録日 | 2020-04-06 | | 公開日 | 2020-10-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Selective N-Terminal BET Bromodomain Inhibitors by Targeting Non-Conserved Residues and Structured Water Displacement*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
8HXX
 
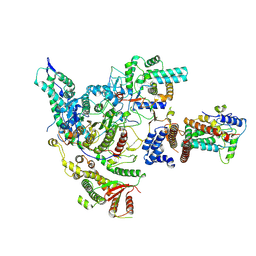 | |
8HXY
 
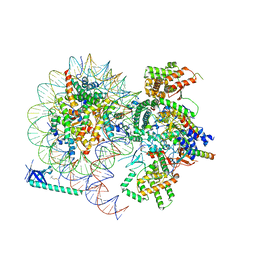 | |
8HY0
 
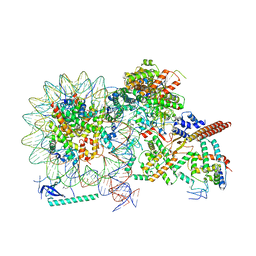 | |
8HXZ
 
 | |
5D8M
 
 | | Crystal structure of the metagenomic carboxyl esterase MGS0156 | | 分子名称: | Metagenomic carboxyl esterase MGS0156 | | 著者 | Cui, H, Nocek, B, Tchigvintsev, A, Popovic, A, Savchenko, A, Joachimiak, A, Yakunin, A. | | 登録日 | 2015-08-17 | | 公開日 | 2016-10-05 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of esterase (MGS0156)
To Be Published
|
|
4FCG
 
 | | Structure of the leucine-rich repeat domain of the type III effector XCV3220 (XopL) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | 著者 | Singer, A.U, Xu, X, Cui, H, Zimmerman, M.D, Minor, W, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2012-05-24 | | 公開日 | 2012-06-13 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of the leucine-rich repeat domain of the type III effector XCV3220 (XopL)
To be Published
|
|
4FC9
 
 | | Structure of the C-terminal domain of the type III effector Xcv3220 (XopL) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, uncharacterized protein | | 著者 | Singer, A.U, Xu, X, Cui, H, Tan, K, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2012-05-24 | | 公開日 | 2012-06-13 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of the C-terminal domain of the type III effector Xcv3220 (XopL)
To be Published
|
|
3TJY
 
 | | Structure of the Pto-binding domain of HopPmaL generated by limited chymotrypsin digestion | | 分子名称: | CHLORIDE ION, Effector protein hopAB3, SULFATE ION | | 著者 | Singer, A.U, Stein, A, Xu, X, Cui, H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-08-25 | | 公開日 | 2011-09-14 | | 最終更新日 | 2013-01-09 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural analysis of HopPmaL reveals the presence of a second adaptor domain common to the HopAB family of Pseudomonas syringae type III effectors.
Biochemistry, 51, 2012
|
|
4RO0
 
 | | Crystal structure of MthK gating ring in a ligand-free form | | 分子名称: | Calcium-gated potassium channel MthK | | 著者 | Dong, W, Guo, R, Chai, H, Chen, Z, Cui, H, Ren, Z, Li, Y, Ye, S. | | 登録日 | 2014-10-27 | | 公開日 | 2015-10-28 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.18 Å) | | 主引用文献 | The principle of cooperative gating in a calcium-gated K+ channel
To be Published
|
|
4MWA
 
 | | 1.85 Angstrom Crystal Structure of GCPE Protein from Bacillus anthracis | | 分子名称: | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, CHLORIDE ION, SULFATE ION | | 著者 | Minasov, G, Wawrzak, Z, Brunzelle, J.S, Xu, X, Cui, H, Maltseva, N, Bishop, B, Kwon, K, Savchenko, A, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-09-24 | | 公開日 | 2013-10-09 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | 1.85 Angstrom Crystal Structure of GCPE Protein from Bacillus anthracis.
TO BE PUBLISHED
|
|
3KWP
 
 | | Crystal structure of putative methyltransferase from Lactobacillus brevis | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Predicted methyltransferase | | 著者 | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-12-01 | | 公開日 | 2009-12-15 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Crystal structure of putative methyltransferase from Lactobacillus brevis
To be Published
|
|
