7PG0
 
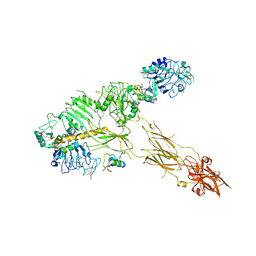 | | Low resolution Cryo-EM structure of full-length insulin receptor bound to 3 insulin with visible ddm micelle, conf 1 | | 分子名称: | Insulin, Isoform Short of Insulin receptor | | 著者 | Nielsen, J.A, Slaaby, R, Boesen, T, Hummelshoj, T, Brandt, J, Schluckebier, G, Nissen, P. | | 登録日 | 2021-08-12 | | 公開日 | 2022-02-02 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (7.6 Å) | | 主引用文献 | Structural Investigations of Full-Length Insulin Receptor Dynamics and Signalling.
J.Mol.Biol., 434, 2022
|
|
7PG2
 
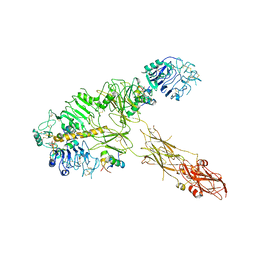 | | Low resolution Cryo-EM structure of full-length insulin receptor bound to 3 insulin, conf 1 | | 分子名称: | Insulin, Isoform Short of Insulin receptor | | 著者 | Nielsen, J.A, Slaaby, R, Boesen, T, Hummelshoj, T, Brandt, J, Schluckebier, G, Nissen, P. | | 登録日 | 2021-08-12 | | 公開日 | 2022-02-02 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (6.7 Å) | | 主引用文献 | Structural Investigations of Full-Length Insulin Receptor Dynamics and Signalling.
J.Mol.Biol., 434, 2022
|
|
7PG4
 
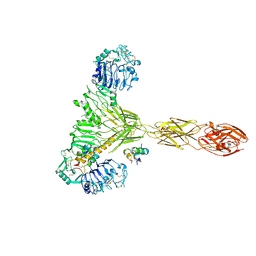 | | Low resolution Cryo-EM structure of the full-length insulin receptor bound to 2 insulin, conf 3 | | 分子名称: | Insulin, Isoform Short of Insulin receptor | | 著者 | Nielsen, J.A, Slaaby, R, Boesen, T, Hummelshoj, T, Brandt, J, Schluckebier, G, Nissen, P. | | 登録日 | 2021-08-12 | | 公開日 | 2022-02-02 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (9.1 Å) | | 主引用文献 | Structural Investigations of Full-Length Insulin Receptor Dynamics and Signalling.
J.Mol.Biol., 434, 2022
|
|
7PG3
 
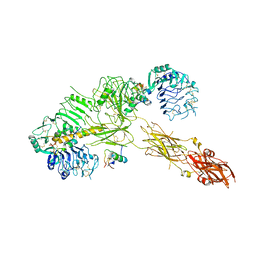 | | Low resolution Cryo-EM structure of the full-length insulin receptor bound to 3 insulin, conf 2 | | 分子名称: | Insulin, Isoform Short of Insulin receptor | | 著者 | Nielsen, J.A, Slaaby, R, Boesen, T, Hummelshoj, T, Brandt, J, Schluckebier, G, Nissen, P. | | 登録日 | 2021-08-12 | | 公開日 | 2022-02-02 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (7.3 Å) | | 主引用文献 | Structural Investigations of Full-Length Insulin Receptor Dynamics and Signalling.
J.Mol.Biol., 434, 2022
|
|
2W44
 
 | | Structure DeltaA1-A4 insulin | | 分子名称: | CHLORIDE ION, INSULIN, RESORCINOL, ... | | 著者 | Thorsoee, K.S, Schlein, M, Brandt, J, Schluckebier, G, Naver, H. | | 登録日 | 2008-11-21 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Kinetic Evidence for the Sequential Association of Insulin Binding Sites 1 and 2 to the Insulin Receptor and the Influence of Receptor Isoform.
Biochemistry, 49, 2010
|
|
5ILC
 
 | | The X-ray structure of the adduct formed in the reaction between hen egg white lysozyme a compound 2, a platin(II) compound containing a O, S bidentate ligand | | 分子名称: | ACETATE ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Merlino, A, Ferraro, G. | | 登録日 | 2016-03-04 | | 公開日 | 2016-12-07 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Platinum(ii) O,S complexes as potential metallodrugs against Cisplatin resistance.
Dalton Trans, 45, 2016
|
|
5ILF
 
 | | The X-ray structure of the adduct formed in the reaction between hen egg white lysozyme and compound 4, a platin(II) compound containing a O, S bidentate ligand | | 分子名称: | ACETATE ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Merlino, A, Ferraro, G. | | 登録日 | 2016-03-04 | | 公開日 | 2016-12-07 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Platinum(ii) O,S complexes as potential metallodrugs against Cisplatin resistance.
Dalton Trans, 45, 2016
|
|
5IHG
 
 | | The X-ray structure of the adduct formed in the reaction between hen egg white lysozyme a compound I, a platin(II) compound containing a O, S bidentate ligand | | 分子名称: | CHLORIDE ION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | 著者 | Ferraro, G, Merlino, A. | | 登録日 | 2016-02-29 | | 公開日 | 2016-12-07 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Platinum(ii) O,S complexes as potential metallodrugs against Cisplatin resistance.
Dalton Trans, 45, 2016
|
|
5JLG
 
 | | The X-ray structure of the adduct formed in the reaction between bovine pancreatic ribonuclease and compound I, a piano-stool organometallic Ru(II) arene compound containing an O,S-chelating ligand | | 分子名称: | DIMETHYL SULFOXIDE, RUTHENIUM ION, Ribonuclease pancreatic, ... | | 著者 | Ferraro, G, Merlino, A. | | 登録日 | 2016-04-27 | | 公開日 | 2016-08-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Unusual mode of protein binding by a cytotoxic pi-arene ruthenium(ii) piano-stool compound containing an O,S-chelating ligand.
Dalton Trans, 45, 2016
|
|
5II3
 
 | | The X-ray structure of the adduct formed in the reaction between hen egg white lysozyme and compound 3, a platin(II) compound containing a O, S bidentate ligand | | 分子名称: | CHLORIDE ION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | 著者 | Ferraro, G, Merlino, A. | | 登録日 | 2016-03-01 | | 公開日 | 2016-12-07 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Platinum(ii) O,S complexes as potential metallodrugs against Cisplatin resistance.
Dalton Trans, 45, 2016
|
|
2WFU
 
 | | Crystal structure of DILP5 variant DB | | 分子名称: | PROBABLE INSULIN-LIKE PEPTIDE 5 A CHAIN, PROBABLE INSULIN-LIKE PEPTIDE 5 B CHAIN | | 著者 | Kulahin, N, Schluckebier, G, Sajid, W, De Meyts, P. | | 登録日 | 2009-04-15 | | 公開日 | 2010-05-26 | | 最終更新日 | 2012-04-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural and Biological Properties of the Drosophila Insulin-Like Peptide 5 Show Evolutionary Conservation.
J.Biol.Chem., 286, 2011
|
|
2WFV
 
 | | Crystal structure of DILP5 variant C4 | | 分子名称: | PROBABLE INSULIN-LIKE PEPTIDE 5 A CHAIN, PROBABLE INSULIN-LIKE PEPTIDE 5 B CHAIN | | 著者 | Kulahin, N, Schluckebier, G, Sajid, W, De Meyts, P. | | 登録日 | 2009-04-15 | | 公開日 | 2010-05-26 | | 最終更新日 | 2012-04-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural and Biological Properties of the Drosophila Insulin-Like Peptide 5 Show Evolutionary Conservation.
J.Biol.Chem., 286, 2011
|
|
8G83
 
 | | Structure of NAD+ consuming protein Acinetobacter baumannii TIR domain | | 分子名称: | NAD(+) hydrolase AbTIR | | 著者 | Klontz, E.H, Wang, Y, Glendening, G, Carr, J, Tsibouris, T, Buddula, S, Nallar, S, Soares, A, Snyder, G.A. | | 登録日 | 2023-02-17 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.03 Å) | | 主引用文献 | The structure of NAD + consuming protein Acinetobacter baumannii TIR domain shows unique kinetics and conformations.
J.Biol.Chem., 299, 2023
|
|
7RTC
 
 | |
2HE7
 
 | | FERM domain of EPB41L3 (DAL-1) | | 分子名称: | Band 4.1-like protein 3 | | 著者 | Hallberg, B.M, Busam, R.D, Arrowsmith, C, Berglund, H, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Schiavone, L.H, Johansson, I, Hogbom, M, Karlberg, T, Kotenyova, T, Nilvebrandt, J, Norberg, P, Stenmark, P, Nordlund, P, Nilsson-ehle, P, Nyman, T, Ogg, D, Sagemark, J, Sundstrom, M, Uppenberg, J, Van den berg, S, Weigelt, J, Persson, C, Thorsell, A.G, Structural Genomics Consortium (SGC) | | 登録日 | 2006-06-21 | | 公開日 | 2006-07-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis of tumor suppressor in lung cancer 1 (TSLC1) binding to differentially expressed in adenocarcinoma of the lung (DAL-1/4.1B).
J.Biol.Chem., 286, 2011
|
|
7KNQ
 
 | | SARM1 Octamer | | 分子名称: | NAD(+) hydrolase SARM1 | | 著者 | Shen, C, Wu, H. | | 登録日 | 2020-11-05 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Multiple domain interfaces mediate SARM1 autoinhibition.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7UWG
 
 | | The crystal structure of the TIR domain-containing protein from Acinetobacter baumannii (AbTir) | | 分子名称: | HEXAETHYLENE GLYCOL, Molecular chaperone Tir, SULFATE ION | | 著者 | Manik, M.K, Nanson, J.D, Ve, T, Kobe, B. | | 登録日 | 2022-05-03 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science, 377, 2022
|
|
7UXR
 
 | | Crystal structure of the BtTir TIR domain | | 分子名称: | TIR domain protein | | 著者 | Shi, Y, Masic, V, Mosaiab, T, Vasquez, E, Ve, T. | | 登録日 | 2022-05-06 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science, 377, 2022
|
|
7UXU
 
 | | CryoEM structure of the TIR domain from AbTir in complex with 3AD | | 分子名称: | Molecular chaperone Tir, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(8-azanylisoquinolin-2-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | 著者 | Li, S, Nanson, J.D, Manik, M.K, Gu, W, Landsberg, M.J, Ve, T, Kobe, B. | | 登録日 | 2022-05-06 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.74 Å) | | 主引用文献 | Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science, 377, 2022
|
|
7UXS
 
 | | Crystal structure of the BcThsA SLOG domain in complex with 3'cADPR | | 分子名称: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, BcThsA, GLYCEROL, ... | | 著者 | Shi, Y, Masic, V, Mosaiab, T, Ve, T. | | 登録日 | 2022-05-06 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science, 377, 2022
|
|
7UXT
 
 | | Crystal structure of ligand-free SeThsA | | 分子名称: | GLYCEROL, TRIETHYLENE GLYCOL, USG protein | | 著者 | Shi, Y, Masic, V, Mosaiab, T, Nanson, J.D, Kobe, B, Ve, T. | | 登録日 | 2022-05-06 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science, 377, 2022
|
|
7LD0
 
 | | Cryo-EM structure of ligand-free Human SARM1 | | 分子名称: | NAD(+) hydrolase SARM1 | | 著者 | Nanson, J.D, Gu, W, Luo, Z, Jia, X, Landsberg, M.J, Kobe, B, Ve, T. | | 登録日 | 2021-01-12 | | 公開日 | 2021-03-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | SARM1 is a metabolic sensor activated by an increased NMN/NAD + ratio to trigger axon degeneration.
Neuron, 109, 2021
|
|
7LCY
 
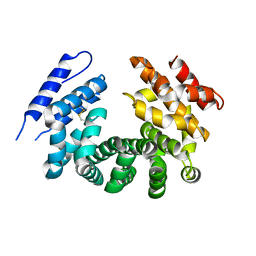 | | Crystal structure of the ligand-free ARM domain from Drosophila SARM1 | | 分子名称: | Isoform B of NAD(+) hydrolase sarm1 | | 著者 | Gu, W, Nanson, J.D, Luo, Z, McGuinness, H.Y, Manik, M.K, Jia, X, Ve, T, Kobe, B. | | 登録日 | 2021-01-12 | | 公開日 | 2021-03-10 | | 最終更新日 | 2021-04-21 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | SARM1 is a metabolic sensor activated by an increased NMN/NAD + ratio to trigger axon degeneration.
Neuron, 109, 2021
|
|
7LCZ
 
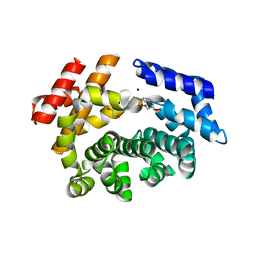 | | Crystal structure of the ARM domain from Drosophila SARM1 in complex with NMN | | 分子名称: | 1,2-ETHANEDIOL, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Isoform B of NAD(+) hydrolase sarm1, ... | | 著者 | Gu, W, Nanson, J.D, Luo, Z, Jia, X, Manik, M.K, Ve, T, Kobe, B. | | 登録日 | 2021-01-12 | | 公開日 | 2021-03-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | SARM1 is a metabolic sensor activated by an increased NMN/NAD + ratio to trigger axon degeneration.
Neuron, 109, 2021
|
|
2GH0
 
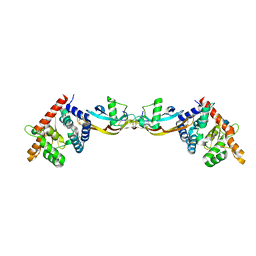 | | Growth factor/receptor complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GDNF family receptor alpha-3, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Wang, X.Q. | | 登録日 | 2006-03-24 | | 公開日 | 2006-06-27 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structure of Artemin Complexed with Its Receptor GFRalpha3: Convergent Recognition of Glial Cell Line-Derived Neurotrophic Factors.
Structure, 14, 2006
|
|
