1A8K
 
 | | CRYSTALLOGRAPHIC ANALYSIS OF HUMAN IMMUNODEFICIENCY VIRUS 1 PROTEASE WITH AN ANALOG OF THE CONSERVED CA-P2 SUBSTRATE: INTERACTIONS WITH FREQUENTLY OCCURRING GLUTAMIC ACID RESIDUE AT P2' POSITION OF SUBSTRATES | | 分子名称: | HIV PROTEASE, N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide | | 著者 | Weber, I.T, Wu, J, Adomat, J, Harrison, R.W, Kimmel, A.R, Wondrak, E.M, Louis, J.M. | | 登録日 | 1998-03-27 | | 公開日 | 1999-01-13 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystallographic analysis of human immunodeficiency virus 1 protease with an analog of the conserved CA-p2 substrate -- interactions with frequently occurring glutamic acid residue at P2' position of substrates.
Eur.J.Biochem., 249, 1997
|
|
4JEC
 
 | | Joint neutron and X-ray structure of per-deuterated HIV-1 protease in complex with clinical inhibitor amprenavir | | 分子名称: | CHLORIDE ION, HIV-1 protease, {3-[(4-AMINO-BENZENESULFONYL)-ISOBUTYL-AMINO]-1-BENZYL-2-HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | 著者 | Kovalevsky, A.Y, Weber, I.T, Langan, P. | | 登録日 | 2013-02-26 | | 公開日 | 2013-07-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | NEUTRON DIFFRACTION (2.01 Å), X-RAY DIFFRACTION | | 主引用文献 | Joint X-ray/Neutron Crystallographic Study of HIV-1 Protease with Clinical Inhibitor Amprenavir: Insights for Drug Design.
J.Med.Chem., 56, 2013
|
|
2HS2
 
 | | Crystal structure of M46L mutant of HIV-1 protease complexed with TMC114 (darunavir) | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Weber, I.T, Kovalevsky, A.Y, Liu, F. | | 登録日 | 2006-07-20 | | 公開日 | 2006-10-03 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | Ultra-high Resolution Crystal Structure of HIV-1 Protease Mutant Reveals Two Binding Sites for Clinical Inhibitor TMC114.
J.Mol.Biol., 363, 2006
|
|
2HS1
 
 | | Ultra-high resolution X-ray crystal structure of HIV-1 protease V32I mutant with TMC114 (darunavir) inhibitor | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Weber, I.T, Kovalevsky, A.Y. | | 登録日 | 2006-07-20 | | 公開日 | 2006-10-03 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (0.84 Å) | | 主引用文献 | Ultra-high Resolution Crystal Structure of HIV-1 Protease Mutant Reveals Two Binding Sites for Clinical Inhibitor TMC114.
J.Mol.Biol., 363, 2006
|
|
3BVA
 
 | | Cystal structure of HIV-1 Active Site Mutant D25N and p2-NC analog inhibitor | | 分子名称: | GLYCEROL, N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, Protease (Retropepsin) | | 著者 | Liu, F, Weber, I.T. | | 登録日 | 2008-01-05 | | 公開日 | 2008-04-01 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Effect of the Active Site D25N Mutation on the Structure, Stability, and Ligand Binding of the Mature HIV-1 Protease.
J.Biol.Chem., 283, 2008
|
|
1FG8
 
 | | STRUCTURAL IMPLICATIONS OF DRUG RESISTANT MUTANTS OF HIV-1 PROTEASE: HIGH RESOLUTION CRYSTAL STRUCTURES OF THE MUTANT PROTEASE/SUBSTRATE ANALOG COMPLEXES | | 分子名称: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE RETROPEPSIN | | 著者 | Mahalingam, B, Louis, J.M, Harrison, R.W, Weber, I.T. | | 登録日 | 2000-07-25 | | 公開日 | 2001-06-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes.
Proteins, 43, 2001
|
|
5JFU
 
 | | HIV-1 wild Type protease with GRL-007-14A (a Adamantane P1-Ligand with bis-THF in P2 and benzylamine in P1') | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-4-{benzyl[(4-methoxyphenyl)sulfonyl]amino}-3-hydroxy-1-[(3R,5R,7R)-tricyclo[3.3.1.1~3,7~]decan-1-yl]butan-2-yl}carbamate, CHLORIDE ION, Protease | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2016-04-19 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Probing Lipophilic Adamantyl Group as the P1-Ligand for HIV-1 Protease Inhibitors: Design, Synthesis, Protein X-ray Structural Studies, and Biological Evaluation.
J.Med.Chem., 59, 2016
|
|
5JG1
 
 | | HIV-1 wild Type protease with GRL-031-14A (a Adamantane P1-Ligand with tetrahydropyrano-tetrahydrofuran in P2 and isobutylamine in P1') | | 分子名称: | (3R,3aS,7aR)-hexahydro-4H-furo[2,3-b]pyran-3-yl {(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-[(3R,5R,7R)-tricyclo[3.3.1.1~3,7~]decan-1-yl]butan-2-yl}carbamate, CHLORIDE ION, GLYCEROL, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2016-04-19 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Probing Lipophilic Adamantyl Group as the P1-Ligand for HIV-1 Protease Inhibitors: Design, Synthesis, Protein X-ray Structural Studies, and Biological Evaluation.
J.Med.Chem., 59, 2016
|
|
4NPT
 
 | | Crystal Structure of HIV-1 Protease Multiple Mutant P51 Complexed with Darunavir | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, GLYCEROL, Protease | | 著者 | Zhang, Y, Weber, I.T. | | 登録日 | 2013-11-22 | | 公開日 | 2014-10-08 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Structures of darunavir-resistant HIV-1 protease mutant reveal atypical binding of darunavir to wide open flaps.
Acs Chem.Biol., 9, 2014
|
|
5JFP
 
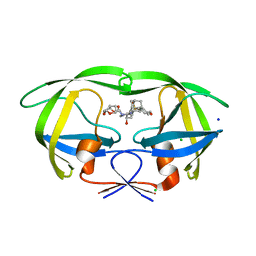 | | HIV-1 wild Type protease with GRL-097-13A (a Adamantane P1-Ligand with bis-THF in P2 and isobutylamine in P1') | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-[(3R,5R,7R)-tricyclo[3.3.1.1~3,7~]decan-1-yl]butan-2-yl}carbamate, CHLORIDE ION, Protease, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2016-04-19 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Probing Lipophilic Adamantyl Group as the P1-Ligand for HIV-1 Protease Inhibitors: Design, Synthesis, Protein X-ray Structural Studies, and Biological Evaluation.
J.Med.Chem., 59, 2016
|
|
4FL8
 
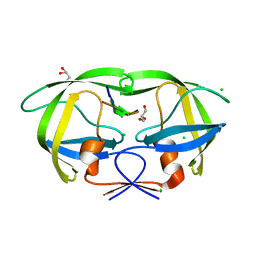 | | HIV-1 protease complexed with gem-diol-amine tetrahedral intermediate | | 分子名称: | CHLORIDE ION, GLYCEROL, HIV-1 protease, ... | | 著者 | Tie, Y.F, Shen, C.H, Weber, I.T. | | 登録日 | 2012-06-14 | | 公開日 | 2012-10-17 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Capturing the Reaction Pathway in Near-Atomic-Resolution Crystal Structures of HIV-1 Protease.
Biochemistry, 51, 2012
|
|
4FLG
 
 | | HIV-1 protease mutant I47V complexed with reaction intermediate | | 分子名称: | CHLORIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | 著者 | Yu, X, Shen, C.H, Weber, I.T. | | 登録日 | 2012-06-14 | | 公開日 | 2012-10-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.31 Å) | | 主引用文献 | Capturing the Reaction Pathway in Near-Atomic-Resolution Crystal Structures of HIV-1 Protease.
Biochemistry, 51, 2012
|
|
3BVB
 
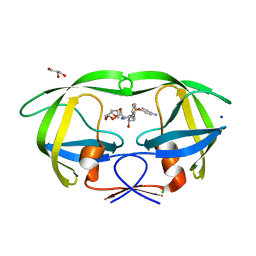 | | Cystal structure of HIV-1 Active Site Mutant D25N and inhibitor Darunavir | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, GLYCEROL, ... | | 著者 | Liu, F, Weber, I.T. | | 登録日 | 2008-01-05 | | 公開日 | 2008-04-01 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Effect of the Active Site D25N Mutation on the Structure, Stability, and Ligand Binding of the Mature HIV-1 Protease.
J.Biol.Chem., 283, 2008
|
|
2AVO
 
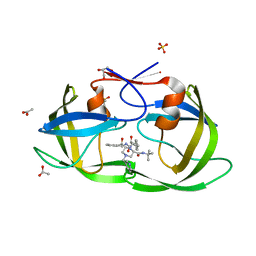 | | Kinetics, stability, and structural changes in high resolution crystal structures of HIV-1 protease with drug resistant mutations L24I, I50V, AND G73S | | 分子名称: | ACETIC ACID, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Liu, F, Boross, P.I, Wang, Y.F, Tozser, J, Louis, J.M, Harrison, R.W, Weber, I.T. | | 登録日 | 2005-08-30 | | 公開日 | 2006-01-24 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Kinetic, stability, and structural changes in high-resolution crystal structures of HIV-1 protease with drug-resistant mutations L24I, I50V, and G73S.
J.Mol.Biol., 354, 2005
|
|
6DJ5
 
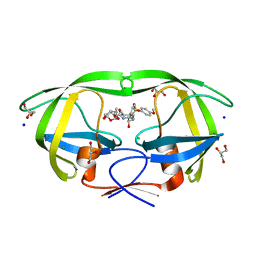 | | HIV-1 protease with mutation L76V in complex with GRL-0519 (tris-tetrahydrofuran as P2 ligand) | | 分子名称: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, GLYCEROL, ... | | 著者 | Wong-Sam, A.E, Wang, Y.F, Weber, I.T. | | 登録日 | 2018-05-24 | | 公開日 | 2018-10-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Drug Resistance Mutation L76V Alters Nonpolar Interactions at the Flap-Core Interface of HIV-1 Protease.
ACS Omega, 3, 2018
|
|
6DV0
 
 | | HIV-1 wild type protease with GRL-02815A, a thiochroman heterocycle with (S)-Boc-amine functionality as the P2 ligand | | 分子名称: | CHLORIDE ION, GLYCEROL, Protease, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2018-06-22 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Design, synthesis, and X-ray studies of potent HIV-1 protease inhibitors incorporating aminothiochromane and aminotetrahydronaphthalene carboxamide derivatives as the P2 ligands.
Eur J Med Chem, 160, 2018
|
|
6E7J
 
 | | HIV-1 wild type protease with GRL-042-17A, 3-phenylhexahydro-2h-cyclopenta[d]oxazol-2-one with a bicyclic oxazolidinone scaffold as the P2 ligand | | 分子名称: | (3aS,5R,6aR)-2-oxo-3-phenylhexahydro-2H-cyclopenta[d][1,3]oxazol-5-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, Protease, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2018-07-26 | | 公開日 | 2018-11-07 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Design and Synthesis of Potent HIV-1 Protease Inhibitors Containing Bicyclic Oxazolidinone Scaffold as the P2 Ligands: Structure-Activity Studies and Biological and X-ray Structural Studies.
J. Med. Chem., 61, 2018
|
|
4U8W
 
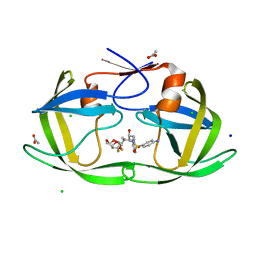 | | HIV-1 wild Type protease with GRL-050-10A (a Gem-difluoro-bis-Tetrahydrofuran as P2-Ligand) | | 分子名称: | (3R,3aS,6aS)-4,4-difluorohexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2014-08-05 | | 公開日 | 2014-11-05 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Design of gem-Difluoro-bis-Tetrahydrofuran as P2 Ligand for HIV-1 Protease Inhibitors to Improve Brain Penetration: Synthesis, X-ray Studies, and Biological Evaluation.
Chemmedchem, 10, 2015
|
|
6O54
 
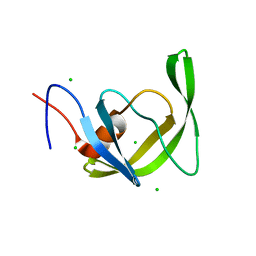 | | Crystal Structure of multi-drug resistant HIV-1 protease PR-S17 (D25N) | | 分子名称: | CHLORIDE ION, HIV-1 protease | | 著者 | Wang, Y.-F, Brothers, R, Agniswamy, J, Weber, I.T. | | 登録日 | 2019-03-01 | | 公開日 | 2019-06-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.21 Å) | | 主引用文献 | Highly Drug-Resistant HIV-1 Protease Mutant PRS17 Shows Enhanced Binding to Substrate Analogues.
Acs Omega, 4, 2019
|
|
6O5X
 
 | | Crystal Structure of multi-drug resistant HIV-1 protease PR-S17 with substrate analog CA-p2 | | 分子名称: | CHLORIDE ION, HIV-1 protease PR-S17, N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, ... | | 著者 | Agniswamy, J, Wang, Y.-F, Weber, I.T. | | 登録日 | 2019-03-04 | | 公開日 | 2019-06-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Highly Drug-Resistant HIV-1 Protease Mutant PRS17 Shows Enhanced Binding to Substrate Analogues.
Acs Omega, 4, 2019
|
|
6O5A
 
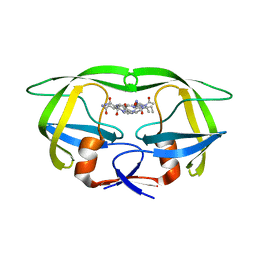 | |
5T2E
 
 | |
5T2Z
 
 | | Crystal Structure of Multi-drug Resistant HIV-1 Protease PR-S17 in Complex with Darunavir | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | 著者 | Agniswamy, J, Weber, I.T. | | 登録日 | 2016-08-24 | | 公開日 | 2017-01-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural Studies of a Rationally Selected Multi-Drug Resistant HIV-1 Protease Reveal Synergistic Effect of Distal Mutations on Flap Dynamics.
PLoS ONE, 11, 2016
|
|
8FUI
 
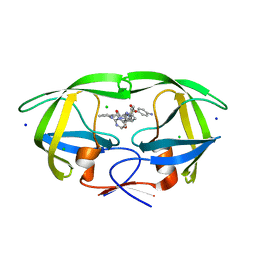 | | HIV-1 wild type protease with GRL-02519A, with N-(2,5-dimethylphenyl)-4-(pyridin-3-yl)pyrimidin-2-amine as P2-P3 group | | 分子名称: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | 著者 | Wang, Y.-F, Wong-Sam, A.E, Ghosh, A.K, Weber, I.T. | | 登録日 | 2023-01-17 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Exploration of imatinib and nilotinib-derived templates as the P2-Ligand for HIV-1 protease inhibitors: Design, synthesis, protein X-ray structural studies, and biological evaluation.
Eur.J.Med.Chem., 255, 2023
|
|
8FUJ
 
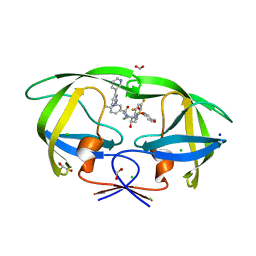 | | HIV-1 wild type protease with GRL-03419A, with N-(2,5-dimethylphenyl)-4-(pyridin-3-yl)pyrimidin-2-amine as P2-P3 group and 3,5-difluorophenylmethyl as the P1 group | | 分子名称: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | 登録日 | 2023-01-17 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | Exploration of imatinib and nilotinib-derived templates as the P2-Ligand for HIV-1 protease inhibitors: Design, synthesis, protein X-ray structural studies, and biological evaluation.
Eur.J.Med.Chem., 255, 2023
|
|
