7XCI
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron RBD in complex with human ACE2 ectodomain (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Gao, G.F, Qi, J.X, Zhao, Z.N, Liu, S, Xie, Y.F. | | 登録日 | 2022-03-24 | | 公開日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7XCP
 
 | | Cryo-EM structure of Omicron RBD complexed with ACE2 and 304 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 304 Fab, ... | | 著者 | Zhao, Z, Qi, J, Gao, F.G. | | 登録日 | 2022-03-24 | | 公開日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7XCO
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with S309 fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 Fab heavy chain, ... | | 著者 | Gao, G.F, Qi, J.X, Zhao, Z.N, Liu, S, Xie, Y.F. | | 登録日 | 2022-03-24 | | 公開日 | 2022-09-21 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
1CB8
 
 | | CHONDROITINASE AC LYASE FROM FLAVOBACTERIUM HEPARINUM | | 分子名称: | CALCIUM ION, GLYCEROL, PROTEIN (CHONDROITINASE AC), ... | | 著者 | Fethiere, J, Eggimann, B, Cygler, M. | | 登録日 | 1999-03-02 | | 公開日 | 1999-05-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of chondroitin AC lyase, a representative of a family of glycosaminoglycan degrading enzymes.
J.Mol.Biol., 288, 1999
|
|
7YA1
 
 | | Cryo-EM structure of hACE2-bound SARS-CoV-2 Omicron spike protein with L371S, P373S and F375S mutations (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | 登録日 | 2022-06-27 | | 公開日 | 2022-08-31 | | 最終更新日 | 2022-09-07 | | 実験手法 | ELECTRON MICROSCOPY (3.11 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7YAD
 
 | | Cryo-EM structure of S309-RBD-RBD-S309 in the S309-bound Omicron spike protein (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, S309 neutralizing antibody heavy chain, S309 neutralizing antibody light chain, ... | | 著者 | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, F. | | 登録日 | 2022-06-27 | | 公開日 | 2022-08-31 | | 最終更新日 | 2022-09-07 | | 実験手法 | ELECTRON MICROSCOPY (2.66 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7Y9S
 
 | | Cryo-EM structure of apo SARS-CoV-2 Omicron spike protein (S-2P-GSAS) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | 登録日 | 2022-06-26 | | 公開日 | 2022-08-31 | | 最終更新日 | 2022-09-07 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7Y9Z
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with human ACE2 ectodomain (one-RBD-up state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Gao, G.F, Qi, J.X, Liu, S, Zhao, Z.N. | | 登録日 | 2022-06-26 | | 公開日 | 2022-09-21 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
8JVB
 
 | | Cryo-EM structure of the Type II secretion system protein from Acidithiobacillus caldus | | 分子名称: | Type II and III secretion system protein | | 著者 | Liu, R.H, Zhang, K, Feng, Q.S, Dai, X, Fu, Y, Li, Y. | | 登録日 | 2023-06-28 | | 公開日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.39 Å) | | 主引用文献 | Cryo-EM structure of the Type II secretion system protein from Acidithiobacillus caldus
To Be Published
|
|
1CJL
 
 | |
3E5H
 
 | | Crystal Structure of Rab28 GTPase in the Active (GppNHp-bound) Form | | 分子名称: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | 著者 | Lee, S.H, Baek, K, Li, Y, Dominguez, R. | | 登録日 | 2008-08-13 | | 公開日 | 2008-08-26 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.499 Å) | | 主引用文献 | Large nucleotide-dependent conformational change in Rab28.
Febs Lett., 582, 2008
|
|
1DIB
 
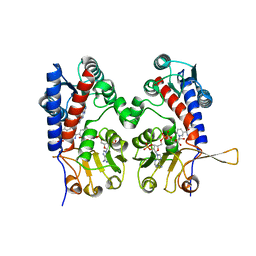 | | HUMAN METHYLENETETRAHYDROFOLATE DEHYDROGENASE / CYCLOHYDROLASE COMPLEXED WITH NADP AND INHIBITOR LY345899 | | 分子名称: | 4-(7-AMINO-9-HYDROXY-1-OXO-3,3A,4,5-TETRAHYDRO-2,5,6,8,9B-PENTAAZA-CYCLOPENTA[A]NAPHTHALEN-2-YL)-PHENYLCARBONYL-GLUTAMI C ACID, METHYLENETETRAHYDROFOLATE DEHYDROGENASE/CYCLOHYDROLASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Schmidt, A, Wu, H, MacKenzie, R.E, Chen, V.J, Bewly, J.R, Ray, J.E, Toth, J.E, Cygler, M. | | 登録日 | 1999-11-29 | | 公開日 | 2000-07-05 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structures of three inhibitor complexes provide insight into the reaction mechanism of the human methylenetetrahydrofolate dehydrogenase/cyclohydrolase.
Biochemistry, 39, 2000
|
|
1DIG
 
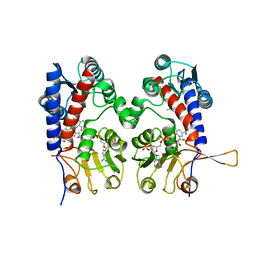 | | HUMAN METHYLENETETRAHYDROFOLATE DEHYDROGENASE / CYCLOHYDROLASE COMPLEXED WITH NADP AND INHIBITOR LY374571 | | 分子名称: | ACETATE ION, METHYLENETETRAHYDROFOLATE DEHYDROGENASE / CYCLOHYDROLASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Schmidt, A, Wu, H, MacKenzie, R.E, Chen, V.J, Bewly, J.R, Ray, J.E, Toth, J.E, Cygler, M. | | 登録日 | 1999-11-29 | | 公開日 | 2000-07-05 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structures of three inhibitor complexes provide insight into the reaction mechanism of the human methylenetetrahydrofolate dehydrogenase/cyclohydrolase.
Biochemistry, 39, 2000
|
|
1DIA
 
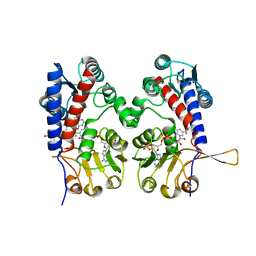 | | HUMAN METHYLENETETRAHYDROFOLATE DEHYDROGENASE / CYCLOHYDROLASE COMPLEXED WITH NADP AND INHIBITOR LY249543 | | 分子名称: | METHYLENETETRAHYDROFOLATE DEHYDROGENASE/CYCLOHYDROLASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [[[2-AMINO-5,6,7,8-TETRAHYDRO-4-HYDROXY-PYRIDO[2,3-D]PYRIMIDIN-6-YL]-ETHYL]-PHENYL]-CARBONYL-GLUTAMIC ACID | | 著者 | Schmidt, A, Wu, H, MacKenzie, R.E, Chen, V.J, Bewly, J.R, Ray, J.E, Toth, J.E, Cygler, M. | | 登録日 | 1999-11-29 | | 公開日 | 2000-07-05 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structures of three inhibitor complexes provide insight into the reaction mechanism of the human methylenetetrahydrofolate dehydrogenase/cyclohydrolase.
Biochemistry, 39, 2000
|
|
7YA0
 
 | | Cryo-EM structure of hACE2-bound SARS-CoV-2 Omicron spike protein with L371S, P373S and F375S mutations (S-6P-RRAR) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | 登録日 | 2022-06-26 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-08-02 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
1DF0
 
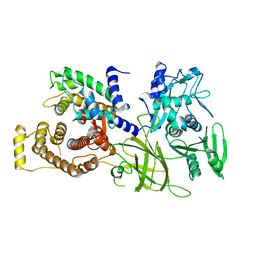 | | Crystal structure of M-Calpain | | 分子名称: | CALPAIN, M-CALPAIN | | 著者 | Hosfield, C.M, Elce, J.S, Davies, P.L, Jia, Z. | | 登録日 | 1999-11-16 | | 公開日 | 2000-06-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of calpain reveals the structural basis for Ca(2+)-dependent protease activity and a novel mode of enzyme activation.
EMBO J., 18, 1999
|
|
3DB3
 
 | | Crystal structure of the tandem tudor domains of the E3 ubiquitin-protein ligase UHRF1 in complex with trimethylated histone H3-K9 peptide | | 分子名称: | E3 ubiquitin-protein ligase UHRF1, Trimethylated histone H3-K9 peptide | | 著者 | Walker, J.R, Avvakumov, G.V, Xue, S, Dong, A, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | 登録日 | 2008-05-30 | | 公開日 | 2008-09-16 | | 最終更新日 | 2012-04-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein.
J.Biol.Chem., 286, 2011
|
|
7EEJ
 
 | | Complex structure of glycoside hydrolase family 12 beta-1,3-1,4-glucanase with cellobiose | | 分子名称: | GLYCEROL, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose, glycoside hydrolase family 12 beta-1,3-1,4-glucanase | | 著者 | Jiang, Z.Q, Ma, J.W. | | 登録日 | 2021-03-18 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.47798049 Å) | | 主引用文献 | Structural and biochemical insights into the substrate-binding mechanism of a glycoside hydrolase family 12 beta-1,3-1,4-glucanase from Chaetomium sp.
J.Struct.Biol., 213, 2021
|
|
7EE2
 
 | | Structural insights into the substrate-binding mechanism of a glycoside hydrolase family 12 beta-1,3-1,4-glucanase from Chaetomium sp.CQ31 | | 分子名称: | GLYCEROL, glycoside hydrolase family 12 beta-1,3-1,4-glucanase | | 著者 | Jiang, Z.Q, Ma, J. | | 登録日 | 2021-03-17 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.37011635 Å) | | 主引用文献 | Structural and biochemical insights into the substrate-binding mechanism of a glycoside hydrolase family 12 beta-1,3-1,4-glucanase from Chaetomium sp.
J.Struct.Biol., 213, 2021
|
|
7EEE
 
 | | Complex structure of glycoside hydrolase family 12 beta-1,3-1,4-glucanase with gentiobiose | | 分子名称: | GLYCEROL, beta-D-mannopyranose-(1-6)-beta-D-mannopyranose, glycoside hydrolase family 12 beta-1,3-1,4-glucanase | | 著者 | Jiang, Z.Q, Ma, J.W. | | 登録日 | 2021-03-18 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.660792 Å) | | 主引用文献 | Structural and biochemical insights into the substrate-binding mechanism of a glycoside hydrolase family 12 beta-1,3-1,4-glucanase from Chaetomium sp.
J.Struct.Biol., 213, 2021
|
|
7ESV
 
 | |
7ESQ
 
 | |
7ESP
 
 | |
7ESO
 
 | |
1R8K
 
 | | PDXA PROTEIN; NAD-DEPENDENT DEHYDROGENASE/CARBOXYLASE; SUBUNIT OF PYRIDOXINE PHOSPHATE BIOSYNTHETIC PROTEIN PDXJ-PDXA [SALMONELLA TYPHIMURIUM] | | 分子名称: | 4-hydroxythreonine-4-phosphate dehydrogenase 1, CHLORIDE ION, COBALT (II) ION, ... | | 著者 | Osipiuk, J, Quartey, P, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2003-10-27 | | 公開日 | 2003-11-11 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of NAD-dependent dehydrogenase/carboxylase of Salmonella typhimurium
to be published
|
|
