8FAK
 
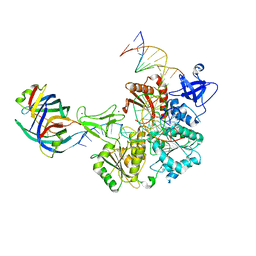 | | DNA replication fork binding triggers structural changes in the PriA DNA helicase that regulate the PriA-PriB replication restart pathway in E. coli | | 分子名称: | DNA (5'-D(P*CP*AP*GP*AP*CP*TP*CP*AP*TP*TP*TP*AP*GP*CP*CP*CP*TP*TP*AP*TP*CP*CP*G)-3'), DNA (5'-D(P*CP*GP*GP*AP*TP*AP*AP*GP*GP*GP*CP*TP*GP*AP*GP*CP*AP*CP*GP*CP*CP*GP*A)-3'), DNA (5'-D(P*TP*CP*GP*GP*CP*GP*TP*GP*CP*TP*C)-3'), ... | | 著者 | Duckworth, A.T, Ducos, P.L, McMillan, S.D, Satyshur, K.A, Blumenthal, K.H, Deorio, H.R, Larson, J.A, Sandler, S.J, Grant, T, Keck, J.L. | | 登録日 | 2022-11-28 | | 公開日 | 2023-05-10 | | 最終更新日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (3.22 Å) | | 主引用文献 | Replication fork binding triggers structural changes in the PriA helicase that govern DNA replication restart in E. coli.
Nat Commun, 14, 2023
|
|
1TL9
 
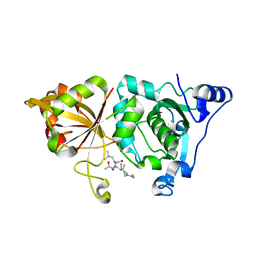 | | High resolution crystal structure of calpain I protease core in complex with leupeptin | | 分子名称: | CALCIUM ION, Calpain 1, large [catalytic] subunit, ... | | 著者 | Moldoveanu, T, Campbell, R.L, Cuerrier, D, Davies, P.L. | | 登録日 | 2004-06-09 | | 公開日 | 2004-11-02 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structures of Calpain-E64 and -Leupeptin Inhibitor Complexes Reveal Mobile Loops Gating the Active Site
J.Mol.Biol., 343, 2004
|
|
1TLO
 
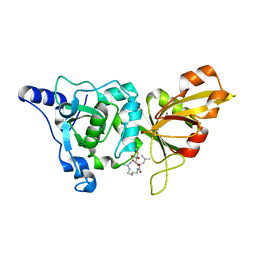 | | High resolution crystal structure of calpain I protease core in complex with E64 | | 分子名称: | CALCIUM ION, Calpain 1, large [catalytic] subunit, ... | | 著者 | Moldoveanu, T, Campbell, R.L, Cuerrier, D, Davies, P.L. | | 登録日 | 2004-06-09 | | 公開日 | 2004-11-02 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structures of Calpain-E64 and -Leupeptin Inhibitor Complexes Reveal Mobile Loops Gating the Active Site
J.Mol.Biol., 343, 2004
|
|
1A7A
 
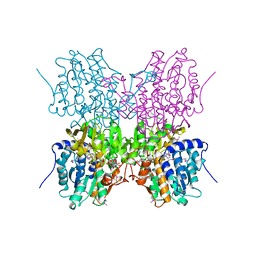 | | STRUCTURE OF HUMAN PLACENTAL S-ADENOSYLHOMOCYSTEINE HYDROLASE: DETERMINATION OF A 30 SELENIUM ATOM SUBSTRUCTURE FROM DATA AT A SINGLE WAVELENGTH | | 分子名称: | (1'R,2'S)-9-(2-HYDROXY-3'-KETO-CYCLOPENTEN-1-YL)ADENINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, S-ADENOSYLHOMOCYSTEINE HYDROLASE | | 著者 | Turner, M.A, Yuan, C.-S, Borchardt, R.T, Hershfield, M.S, Smith, G.D, Howell, P.L. | | 登録日 | 1998-03-10 | | 公開日 | 1999-04-20 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure determination of selenomethionyl S-adenosylhomocysteine hydrolase using data at a single wavelength.
Nat.Struct.Biol., 5, 1998
|
|
1IPS
 
 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (MANGANESE COMPLEX) | | 分子名称: | ISOPENICILLIN N SYNTHASE, MANGANESE (II) ION | | 著者 | Roach, P.L, Clifton, I.J, Fulop, V, Harlos, K, Barton, G.J, Hajdu, J, Andersson, I, Schofield, C.J, Baldwin, J.E. | | 登録日 | 1997-03-21 | | 公開日 | 1998-03-25 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of isopenicillin N synthase is the first from a new structural family of enzymes.
Nature, 375, 1995
|
|
6AU1
 
 | | Structure of the PgaB (BpsB) glycoside hydrolase domain from Bordetella bronchiseptica | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative hemin storage protein, ... | | 著者 | Little, D.J, Bamford, N.C, Howell, P.L. | | 登録日 | 2017-08-30 | | 公開日 | 2018-04-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | PgaB orthologues contain a glycoside hydrolase domain that cleaves deacetylated poly-beta (1,6)-N-acetylglucosamine and can disrupt bacterial biofilms.
PLoS Pathog., 14, 2018
|
|
6BDT
 
 | |
6BG8
 
 | |
6BJD
 
 | | Crystal Structure of Human Calpain-3 Protease Core in Complex with E-64 | | 分子名称: | CALCIUM ION, CHLORIDE ION, Calpain-3, ... | | 著者 | Ye, Q, Campbell, R.L, Davies, P.L. | | 登録日 | 2017-11-06 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structures of human calpain-3 protease core with and without bound inhibitor reveal mechanisms of calpain activation.
J. Biol. Chem., 293, 2018
|
|
6BGP
 
 | |
6BKJ
 
 | |
7ULA
 
 | | Structure of the Pseudomonas putida AlgKX modification and secretion complex | | 分子名称: | Alginate biosynthesis protein AlgK, Alginate biosynthesis protein AlgX, CHLORIDE ION, ... | | 著者 | Gheorghita, A.A, Li, E.Y, Pfoh, R, Howell, P.L. | | 登録日 | 2022-04-04 | | 公開日 | 2022-12-14 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Structure of the AlgKX modification and secretion complex required for alginate production and biofilm attachment in Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
6BT4
 
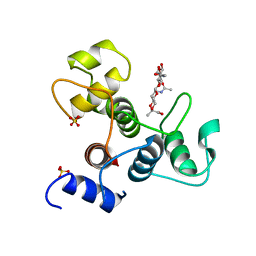 | | Crystal structure of the SLH domain of Sap from Bacillus anthracis in complex with a pyruvylated SCWP unit | | 分子名称: | 2-(acetylamino)-4-O-{2-(acetylamino)-4,6-O-[(1S)-1-carboxyethylidene]-2-deoxy-beta-D-mannopyranosyl}-2-deoxy-beta-D-glucopyranose, S-layer protein sap, SULFATE ION | | 著者 | Sychantha, D, Chapman, R.N, Bamford, N.C, Boons, G.J, Howell, P.L, Clarke, A.J. | | 登録日 | 2017-12-05 | | 公開日 | 2018-03-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.306 Å) | | 主引用文献 | Molecular Basis for the Attachment of S-Layer Proteins to the Cell Wall of Bacillus anthracis.
Biochemistry, 57, 2018
|
|
1B3C
 
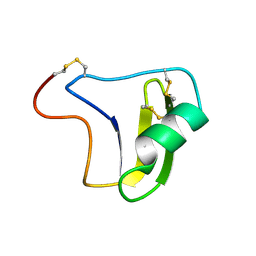 | | SOLUTION STRUCTURE OF A BETA-NEUROTOXIN FROM THE NEW WORLD SCORPION CENTRUROIDES SCULPTURATUS EWING | | 分子名称: | PROTEIN (NEUROTOXIN CSE-I) | | 著者 | Jablonsky, M.J, Jackson, P.L, Trent, J.O, Watt, D.D, Krishna, N.R. | | 登録日 | 1998-12-08 | | 公開日 | 1998-12-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a beta-neurotoxin from the New World scorpion Centruroides sculpturatus Ewing.
Biochem.Biophys.Res.Commun., 254, 1999
|
|
1AUW
 
 | |
1AOS
 
 | |
6LA8
 
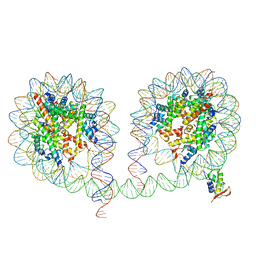 | | 349 bp di-nucleosome harboring cohesive DNA termini assembled with linker histone H1.0 | | 分子名称: | CALCIUM ION, DNA (349-MER), Histone H1.0, ... | | 著者 | Adhireksan, Z, Lee, P.L, Sharma, D, Davey, C.A. | | 登録日 | 2019-11-12 | | 公開日 | 2020-10-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Near-atomic resolution structures of interdigitated nucleosome fibres.
Nat Commun, 11, 2020
|
|
1C55
 
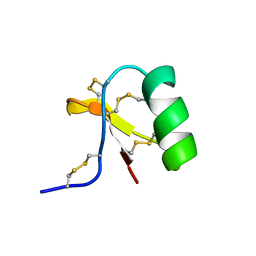 | |
6CHG
 
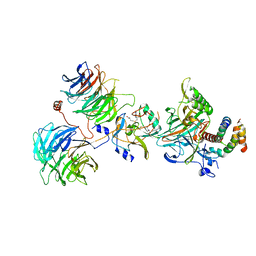 | | Crystal structure of the yeast COMPASS catalytic module | | 分子名称: | H3, Histone-lysine N-methyltransferase, H3 lysine-4 specific, ... | | 著者 | Hsu, P.L, Li, H, Zheng, N. | | 登録日 | 2018-02-22 | | 公開日 | 2018-08-22 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.985 Å) | | 主引用文献 | Crystal Structure of the COMPASS H3K4 Methyltransferase Catalytic Module.
Cell, 174, 2018
|
|
6LA9
 
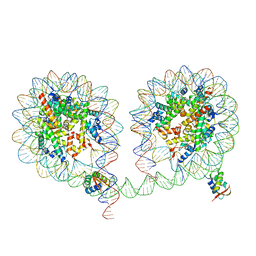 | | 349 bp di-nucleosome harboring cohesive DNA termini assembled with linker histone H1.0 (high cryoprotectant) | | 分子名称: | CALCIUM ION, DNA (349-MER), Histone H1.0, ... | | 著者 | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | 登録日 | 2019-11-12 | | 公開日 | 2020-10-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Near-atomic resolution structures of interdigitated nucleosome fibres.
Nat Commun, 11, 2020
|
|
6CZT
 
 | |
1C56
 
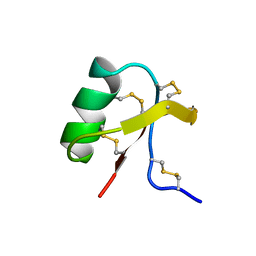 | |
6L9Z
 
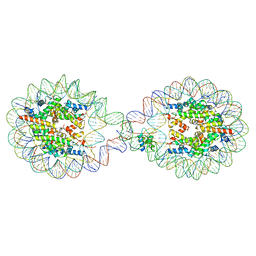 | | 338 bp di-nucleosome assembled with linker histone H1.X | | 分子名称: | CALCIUM ION, CHLORIDE ION, DNA (338-MER), ... | | 著者 | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | 登録日 | 2019-11-11 | | 公開日 | 2021-02-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
6LAB
 
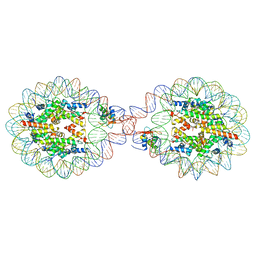 | | 169 bp nucleosome, harboring cohesive DNA termini, assembled with linker histone H1.0 | | 分子名称: | CALCIUM ION, CHLORIDE ION, DNA (169-MER), ... | | 著者 | Adhireksan, Z, Sharma, D, Bao, Q, Lee, P.L, Padavattan, S, Davey, C.A. | | 登録日 | 2019-11-12 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
6LA2
 
 | | 343 bp di-nucleosome harboring cohesive DNA termini assembled with linker histone H1.0 | | 分子名称: | DNA (343-MER), Histone H1.0, Histone H2A type 1-B/E, ... | | 著者 | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | 登録日 | 2019-11-11 | | 公開日 | 2021-02-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.89 Å) | | 主引用文献 | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
