8HO0
 
 | | Crystal structure of cytochrome P450 NasF5053 mutant E73S complexed with 8FCWP | | 分子名称: | (3~{S},8~{a}~{S})-3-[(7-fluoranyl-1~{H}-indol-3-yl)methyl]-2,3,6,7,8,8~{a}-hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, CALCIUM ION, Cytochrome P450-F5053, ... | | 著者 | Ma, B.D, Tian, W, Qu, X, Kong, X.D. | | 登録日 | 2022-12-09 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Engineering the Substrate Specificity of a P450 Dimerase Enables the Collective Biosynthesis of Heterodimeric Tryptophan-Containing Diketopiperazines.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HNY
 
 | | Crystal structure of cytochrome P450 NasF5053 mutant E73S complexed with 5FCWP | | 分子名称: | (3~{S},8~{a}~{S})-3-[(4-fluoranyl-1~{H}-indol-3-yl)methyl]-2,3,6,7,8,8~{a}-hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, Cytochrome P450-F5053, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Ma, B.D, Tian, W, Qu, X, Kong, X.D. | | 登録日 | 2022-12-09 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Engineering the Substrate Specificity of a P450 Dimerase Enables the Collective Biosynthesis of Heterodimeric Tryptophan-Containing Diketopiperazines.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HFK
 
 | |
8HFJ
 
 | | Crystal Structure of CbAR mutant (H162F) in complex with NADP+ and a bulky 1,3-cyclodiketone | | 分子名称: | 2-methyl-2-[(4-methylphenyl)methyl]cyclopentane-1,3-dione, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Versicolorin reductase | | 著者 | Hou, X.D, Yin, D.J, Rao, Y.J. | | 登録日 | 2022-11-10 | | 公開日 | 2023-06-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural analysis of an anthrol reductase inspires enantioselective synthesis of enantiopure hydroxycycloketones and beta-halohydrins.
Nat Commun, 14, 2023
|
|
8H3T
 
 | | The crystal structure of AlpH | | 分子名称: | AlpH, GLYCEROL | | 著者 | Zhao, Y, Li, M, Jiang, M, Pan, L.F. | | 登録日 | 2022-10-09 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.866 Å) | | 主引用文献 | O-methyltransferase-like enzyme catalyzed diazo installation in polyketide biosynthesis.
Nat Commun, 14, 2023
|
|
8IS2
 
 | |
7FH5
 
 | | Structure of AdaV | | 分子名称: | AdaV, CHLORIDE ION, FE (III) ION | | 著者 | Zhang, Z.Y, Chen, W.Q, Zhai, G.Q, Zhang, M. | | 登録日 | 2021-07-29 | | 公開日 | 2022-08-03 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Insight into the Catalytic Mechanism of Non-Heme Iron Halogenase AdaV in 2'-Chloropentostatin Biosynthesis
Acs Catalysis, 12, 2022
|
|
8VY9
 
 | | CryoEM structure of Ggust-coupled TAS2R14 with cholesterol and an intracellular tastant | | 分子名称: | 4-methyl-N-[(2M)-2-(1H-tetrazol-5-yl)phenyl]-6-(trifluoromethyl)pyrimidin-2-amine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Kim, Y, Gumpper, R.H, Roth, B.L. | | 登録日 | 2024-02-07 | | 公開日 | 2024-04-03 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.88 Å) | | 主引用文献 | Bitter taste receptor activation by cholesterol and an intracellular tastant.
Nature, 628, 2024
|
|
8WRJ
 
 | | glycosyltransferase UGT74AN3 | | 分子名称: | 5-[(1R,2S,4R,6R,7R,10S,11S,14S,16R)-14-hydroxy-7,11-dimethyl-3-oxapentacyclo[8.8.0.02,4.02,7.011,16]octadecan-6-yl]pyran-2-one, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | 著者 | Huang, W, Long, F. | | 登録日 | 2023-10-15 | | 公開日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
8WRK
 
 | | glycosyltransferase UGT74AN3 | | 分子名称: | Glycosyltransferase, PICEATANNOL, URIDINE-5'-DIPHOSPHATE | | 著者 | Huang, W, Long, F. | | 登録日 | 2023-10-15 | | 公開日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
8XON
 
 | |
8XOP
 
 | |
8XOO
 
 | |
8XN4
 
 | |
8VY7
 
 | | CryoEM structure of Gi-coupled TAS2R14 with cholesterol and an intracellular tastant | | 分子名称: | 4-methyl-N-[(2M)-2-(1H-tetrazol-5-yl)phenyl]-6-(trifluoromethyl)pyrimidin-2-amine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Kim, Y, Gumpper, R.H, Roth, B.L. | | 登録日 | 2024-02-07 | | 公開日 | 2024-04-03 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (2.68 Å) | | 主引用文献 | Bitter taste receptor activation by cholesterol and an intracellular tastant.
Nature, 628, 2024
|
|
7WVZ
 
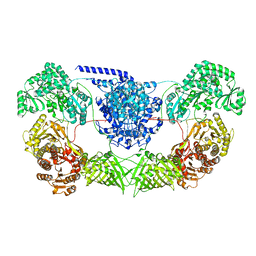 | | CalA3_modular PKS_KS-AT-DH-KR | | 分子名称: | Beta-ketoacyl-acyl-carrier-protein synthase I | | 著者 | Wang, J, Wang, Z. | | 登録日 | 2022-02-12 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3.38 Å) | | 主引用文献 | C-N bond formation by a polyketide synthase.
Nat Commun, 14, 2023
|
|
7Y3Y
 
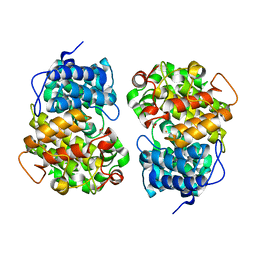 | | Crystal structure of BTG13 mutant (T299V) | | 分子名称: | FE (III) ION, GLYCEROL, questin oxidase BTG13 | | 著者 | Hou, X.D, Fu, K, Rao, Y.J. | | 登録日 | 2022-06-13 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Discovery of the Biosynthetic Pathway of Beticolin 1 Reveals a Novel Non-Heme Iron-Dependent Oxygenase for Anthraquinone Ring Cleavage.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7Y3X
 
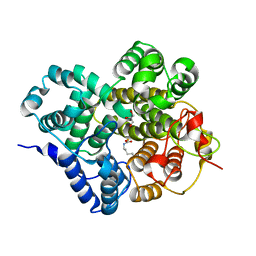 | | Crystal structure of BTG13 mutant (H58F) | | 分子名称: | FE (III) ION, GLYCEROL, Questin oxidase | | 著者 | Hou, X.D, Rao, Y.J. | | 登録日 | 2022-06-13 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Discovery of the Biosynthetic Pathway of Beticolin 1 Reveals a Novel Non-Heme Iron-Dependent Oxygenase for Anthraquinone Ring Cleavage.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7Y3W
 
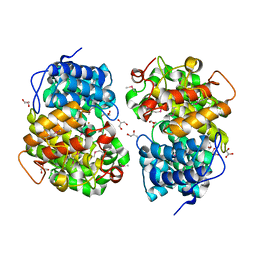 | |
7CPY
 
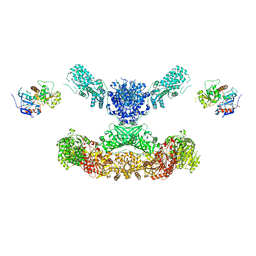 | | Lovastatin nonaketide synthase with LovC | | 分子名称: | Lovastatin nonaketide synthase, enoyl reductase component lovC, polyketide synthase component, ... | | 著者 | Wang, J, Wang, Z. | | 登録日 | 2020-08-08 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural basis for the biosynthesis of lovastatin.
Nat Commun, 12, 2021
|
|
7CPX
 
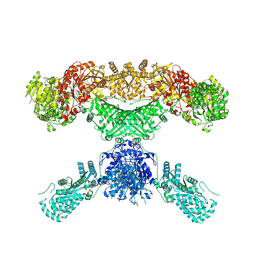 | | Lovastatin nonaketide synthase | | 分子名称: | Lovastatin nonaketide synthase, polyketide synthase component, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Wang, J, Wang, Z. | | 登録日 | 2020-08-08 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.91 Å) | | 主引用文献 | Structural basis for the biosynthesis of lovastatin.
Nat Commun, 12, 2021
|
|
8H0L
 
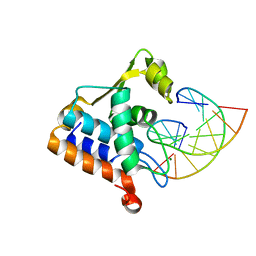 | | Sulfur binding domain of Hga complexed with phosphorothioated DNA | | 分子名称: | DNA (5'-D(*CP*GP*AP*GP*(PST)P*TP*CP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*GP*AP*AP*CP*TP*CP*G)-3'), MAGNESIUM ION, ... | | 著者 | Liu, G, He, X, Hu, W, Yang, B, Xiao, Q. | | 登録日 | 2022-09-29 | | 公開日 | 2023-09-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Characterization of a promiscuous DNA sulfur binding domain and application in site-directed RNA base editing.
Nucleic Acids Res., 51, 2023
|
|
8HVR
 
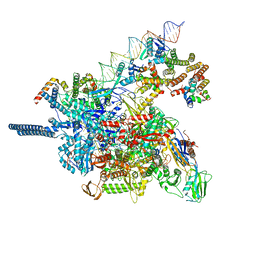 | |
8IND
 
 | | Crystal structure of UGT74AN3-UDP-RES | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-[(1R,2S,4R,6R,7R,10S,11S,14S,16R)-14-hydroxy-7,11-dimethyl-3-oxapentacyclo[8.8.0.02,4.02,7.011,16]octadecan-6-yl]pyran-2-one, Glycosyltransferase, ... | | 著者 | Huang, W. | | 登録日 | 2023-03-09 | | 公開日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
8INV
 
 | | Crystal structure of UGT74AN3-UDP-BUF | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE, ... | | 著者 | Long, F, Huang, W. | | 登録日 | 2023-03-10 | | 公開日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
