5XYB
 
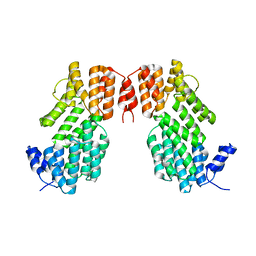 | |
5Y24
 
 | | Crystal structure of AimR from Bacillus phage SPbeta in complex with its signalling peptide | | 分子名称: | AimR transcriptional regulator, BROMIDE ION, GLY-MET-PRO-ARG-GLY-ALA | | 著者 | Wang, Q, Guan, Z.Y, Zou, T.T, Yin, P. | | 登録日 | 2017-07-24 | | 公開日 | 2018-09-19 | | 最終更新日 | 2018-11-28 | | 実験手法 | X-RAY DIFFRACTION (1.922 Å) | | 主引用文献 | Structural basis of the arbitrium peptide-AimR communication system in the phage lysis-lysogeny decision.
Nat Microbiol, 3, 2018
|
|
3QUX
 
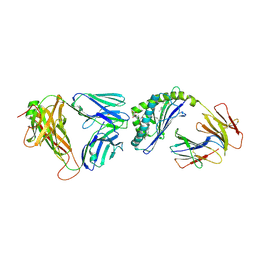 | | Structure of the mouse CD1d-alpha-C-GalCer-iNKT TCR complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | 著者 | Li, Y, Girardi, E, Yu, E.D, Zajonc, D.M. | | 登録日 | 2011-02-24 | | 公開日 | 2011-06-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.91 Å) | | 主引用文献 | Galactose-modified iNKT cell agonists stabilized by an induced fit of CD1d prevent tumour metastasis.
Embo J., 30, 2011
|
|
3QE7
 
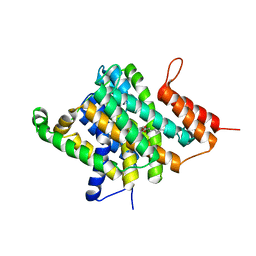 | |
3QUZ
 
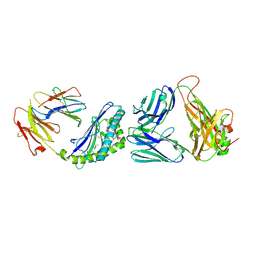 | | Structure of the mouse CD1d-NU-alpha-GalCer-iNKT TCR complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | 著者 | Li, Y, Girardi, E, Yu, E.D, Zajonc, D.M. | | 登録日 | 2011-02-24 | | 公開日 | 2011-06-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Galactose-modified iNKT cell agonists stabilized by an induced fit of CD1d prevent tumour metastasis.
Embo J., 30, 2011
|
|
6K4W
 
 | | smart chimeric peptide SCP-A6 | | 分子名称: | SCP-A6 | | 著者 | Wang, J.H, Liu, X.H. | | 登録日 | 2019-05-27 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Development of chimeric peptides to facilitate the neutralisation of lipopolysaccharides during bactericidal targeting of multidrug-resistant Escherichia coli.
Commun Biol, 3, 2020
|
|
2K3K
 
 | |
5TH7
 
 | | Complex of SETD8 with MS453 | | 分子名称: | 1,2-ETHANEDIOL, N-(3-{[6,7-dimethoxy-2-(pyrrolidin-1-yl)quinazolin-4-yl]amino}propyl)propanamide, N-lysine methyltransferase KMT5A, ... | | 著者 | Yu, W, Tempel, W, Babault, N, Ma, A, Butler, K.V, Jin, J, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | 登録日 | 2016-09-29 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure-Based Design of a Covalent Inhibitor of the SET Domain-Containing Protein 8 (SETD8) Lysine Methyltransferase.
J. Med. Chem., 59, 2016
|
|
2Q06
 
 | | Crystal structure of Influenza A Virus H5N1 Nucleoprotein | | 分子名称: | Nucleoprotein | | 著者 | Ng, A.K.L, Zhang, H, Tan, K, Wang, J, Shaw, P.C. | | 登録日 | 2007-05-21 | | 公開日 | 2008-05-27 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structure of the influenza virus A H5N1 nucleoprotein: implications for RNA binding, oligomerization, and vaccine design.
Faseb J., 22, 2008
|
|
7F65
 
 | | Bacetrial Cocaine Esterase with mutations T172R/G173Q/V116K/S117A/A51L, bound to benzoic acid | | 分子名称: | BENZOIC ACID, Cocaine esterase, SULFATE ION | | 著者 | Ouyang, P.F, Zhang, Y, Tong, J. | | 登録日 | 2021-06-24 | | 公開日 | 2021-09-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.202 Å) | | 主引用文献 | Computational Design and Crystal Structure of a Highly Efficient Benzoylecgonine Hydrolase.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7CSZ
 
 | | Crystal structure of the N-terminal tandem RRM domains of RBM45 in complex with single-stranded DNA | | 分子名称: | DNA (5'-D(*CP*GP*AP*CP*GP*GP*GP*AP*CP*GP*C)-3'), RNA-binding protein 45 | | 著者 | Chen, X, Yang, Z, Wang, W, Wang, M. | | 登録日 | 2020-08-17 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for RNA recognition by the N-terminal tandem RRM domains of human RBM45.
Nucleic Acids Res., 49, 2021
|
|
8TZX
 
 | | Ternary complex structure of Cereblon-DDB1 bound to WIZ(ZF7) and the molecular glue dWIZ-1 | | 分子名称: | (3S)-3-(5-{(1R)-1-[(2R)-1-ethylpiperidin-2-yl]ethoxy}-1-oxo-1,3-dihydro-2H-isoindol-2-yl)piperidine-2,6-dione, 1,2-ETHANEDIOL, DNA damage-binding protein 1, ... | | 著者 | Clifton, M.C, Ma, X, Ornelas, E. | | 登録日 | 2023-08-28 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-07-17 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | A molecular glue degrader of the WIZ transcription factor for fetal hemoglobin induction.
Science, 385, 2024
|
|
7CSX
 
 | |
7CKG
 
 | | Crystal structure of TMSiPheRS complexed with TMSiPhe | | 分子名称: | 4-(trimethylsilyl)-L-phenylalanine, Tyrosine--tRNA ligase | | 著者 | Sun, J.P, Wang, J.Y, Zhu, Z.L, He, Q.T, Xiao, P. | | 登録日 | 2020-07-17 | | 公開日 | 2021-03-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.053 Å) | | 主引用文献 | DeSiphering receptor core-induced and ligand-dependent conformational changes in arrestin via genetic encoded trimethylsilyl 1 H-NMR probe.
Nat Commun, 11, 2020
|
|
7CKH
 
 | | Crystal structure of TMSiPheRS | | 分子名称: | Tyrosine--tRNA ligase | | 著者 | Sun, J.P, Wang, J.Y, Zhu, Z.L, He, Q.T, Xiao, P. | | 登録日 | 2020-07-17 | | 公開日 | 2021-03-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.79492676 Å) | | 主引用文献 | DeSiphering receptor core-induced and ligand-dependent conformational changes in arrestin via genetic encoded trimethylsilyl 1 H-NMR probe.
Nat Commun, 11, 2020
|
|
4PDB
 
 | |
6K0R
 
 | | Ruvbl1-Ruvbl2 with truncated domain II in complex with phosphorylated Cordycepin | | 分子名称: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Zhang, W, Chen, L, Li, W, Ju, D, Huang, N, Zhang, E. | | 登録日 | 2019-05-07 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.502 Å) | | 主引用文献 | Chemical perturbations reveal that RUVBL2 regulates the circadian phase in mammals.
Sci Transl Med, 12, 2020
|
|
3UDC
 
 | | Crystal structure of a membrane protein | | 分子名称: | Small-conductance mechanosensitive channel, C-terminal peptide from Small-conductance mechanosensitive channel | | 著者 | Li, W, Ge, J, Yang, M. | | 登録日 | 2011-10-28 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.355 Å) | | 主引用文献 | Structure and molecular mechanism of an anion-selective mechanosensitive channel of small conductance
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6KRG
 
 | | Crystal structure of sfGFP Y182TMSiPhe | | 分子名称: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | 著者 | Sun, J.P, Wang, J.Y, Zhu, Z.L, He, Q.T, Xiao, P. | | 登録日 | 2019-08-21 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | DeSiphering receptor core-induced and ligand-dependent conformational changes in arrestin via genetic encoded trimethylsilyl 1 H-NMR probe.
Nat Commun, 11, 2020
|
|
6AEI
 
 | | Cryo-EM structure of the receptor-activated TRPC5 ion channel | | 分子名称: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, CHOLESTEROL HEMISUCCINATE, SODIUM ION, ... | | 著者 | Duan, J, Li, Z, Li, J, Zhang, J. | | 登録日 | 2018-08-05 | | 公開日 | 2019-08-07 | | 最終更新日 | 2019-08-14 | | 実験手法 | ELECTRON MICROSCOPY (2.89 Å) | | 主引用文献 | Cryo-EM structure of TRPC5 at 2.8- angstrom resolution reveals unique and conserved structural elements essential for channel function.
Sci Adv, 5, 2019
|
|
5VND
 
 | | Crystal structure of FGFR1-Y563C (FGFR4 surrogate) covalently bound to H3B-6527 | | 分子名称: | 1,2-ETHANEDIOL, Fibroblast growth factor receptor 1, N-{2-[(6-{[(2,6-dichloro-3,5-dimethoxyphenyl)carbamoyl](methyl)amino}pyrimidin-4-yl)amino]-5-(4-ethylpiperazin-1-yl)phenyl}propanamide, ... | | 著者 | Tsai, J.H.C, Reynolds, D, Fekkes, P, Smith, P, Larsen, N.A. | | 登録日 | 2017-04-30 | | 公開日 | 2017-05-24 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | H3B-6527 Is a Potent and Selective Inhibitor of FGFR4 in FGF19-Driven Hepatocellular Carcinoma.
Cancer Res., 77, 2017
|
|
3E88
 
 | | Crystal structures of the kinase domain of AKT2 in complex with ATP-competitive inhibitors | | 分子名称: | 4-[2-(4-amino-1,2,5-oxadiazol-3-yl)-6-{[(2R)-2-amino-3-phenylpropyl]oxy}-1-ethyl-1H-imidazo[4,5-c]pyridin-4-yl]-2-methylbut-3-yn-2-ol, Glycogen synthase kinase-3 beta peptide, RAC-beta serine/threonine-protein kinase | | 著者 | Concha, N.O, Elkins, P.A, Smallwood, A, Ward, P. | | 登録日 | 2008-08-19 | | 公開日 | 2008-10-14 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Aminofurazans as potent inhibitors of AKT kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
7Y01
 
 | |
3E8C
 
 | | Crystal structures of the kinase domain of PKA in complex with ATP-competitive inhibitors | | 分子名称: | 4-[2-(4-amino-1,2,5-oxadiazol-3-yl)-6-{[(2R)-2-amino-3-phenylpropyl]oxy}-1-ethyl-1H-imidazo[4,5-c]pyridin-4-yl]-2-methylbut-3-yn-2-ol, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor peptide | | 著者 | Concha, N.O, Elkins, P.A, Smallwood, A, Ward, P. | | 登録日 | 2008-08-19 | | 公開日 | 2008-11-18 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Aminofurazans as potent inhibitors of AKT kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3E87
 
 | | Crystal structures of the kinase domain of AKT2 in complex with ATP-competitive inhibitors | | 分子名称: | Glycogen synthase kinase-3 beta peptide, N-[(1S)-2-amino-1-phenylethyl]-5-(1H-pyrrolo[2,3-b]pyridin-4-yl)thiophene-2-carboxamide, RAC-beta serine/threonine-protein kinase | | 著者 | Concha, N.O, Elkins, P.A, Smallwood, A, Ward, P. | | 登録日 | 2008-08-19 | | 公開日 | 2008-10-14 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Aminofurazans as potent inhibitors of AKT kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
