1T7J
 
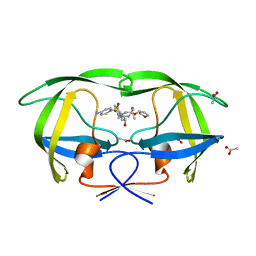 | | crystal structure of inhibitor amprenavir in complex with a multi-drug resistant variant of HIV-1 protease (L63P/V82T/I84V) | | 分子名称: | ACETATE ION, Pol Polyprotein, {3-[(4-AMINO-BENZENESULFONYL)-ISOBUTYL-AMINO]-1-BENZYL-2-HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | 著者 | King, N.M, Prabu-Jeyabalan, M, Nalivaika, E.A, Wigerinck, P.B.T.P, De Bethune, M.-P, Schiffer, C.A. | | 登録日 | 2004-05-10 | | 公開日 | 2005-05-10 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Discovery and selection of TMC114, a next generation HIV-1 protease inhibitor
J.Med.Chem., 48, 2005
|
|
1T3R
 
 | | HIV protease wild-type in complex with TMC114 inhibitor | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, PHOSPHATE ION, protease RETROPEPSIN | | 著者 | King, N.M, Prabu-Jeyabalan, M, Nalivaika, E.A, Wigernick, P.B, de Bethune, M.P, Schiffer, C.A. | | 登録日 | 2004-04-27 | | 公開日 | 2005-05-03 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Discovery and selection of TMC114, a next generation HIV-1 protease inhibitor
J.Med.Chem., 48, 2005
|
|
1T7I
 
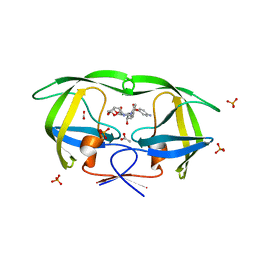 | | The structural and thermodynamic basis for the binding of TMC114, a next-generation HIV-1 protease inhibitor. | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, PHOSPHATE ION, ... | | 著者 | King, N.M, Prabu-Jeyabalan, M, Nalivaika, E.A, Wigerinck, P.B.T.P, De Bethune, M.-P, Schiffer, C.A. | | 登録日 | 2004-05-10 | | 公開日 | 2005-05-10 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Discovery and selection of TMC114, a next generation HIV-1 protease inhibitor
J.Med.Chem., 48, 2005
|
|
1TSU
 
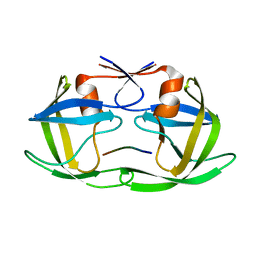 | | CRYSTAL STRUCTURE OF DECAMER NCP1 SUBSTRATE PEPTIDE IN COMPLEX WITH WILD-TYPE D25N HIV-1 PROTEASE VARIANT | | 分子名称: | NC-P1 SUBSTRATE PEPTIDE, Pol polyprotein | | 著者 | Prabu-Jeyabalan, M, Nalivaika, E.A, King, N.M, Schiffer, C.A. | | 登録日 | 2004-06-21 | | 公開日 | 2005-03-29 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for coevolution of a human immunodeficiency virus type 1 nucleocapsid-p1 cleavage site with a V82A drug-resistant mutation in viral protease
J.Virol., 78, 2004
|
|
2FGV
 
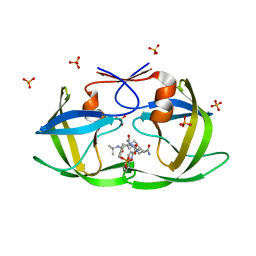 | | X-ray crystal structure of HIV-1 Protease T80N variant in complex with the inhibitor saquinavir used to explore the role of invariant Thr80 in HIV-1 protease structure, function, and viral infectivity. | | 分子名称: | (2S)-2-amino-3-phenylpropane-1,1-diol, 2-METHYL-DECAHYDRO-ISOQUINOLINE-3-CARBOXYLIC ACID, ASPARAGINE, ... | | 著者 | Foulkes, J.E, Prabu-Jeyabalan, M, Cooper, D, Schiffer, C.A. | | 登録日 | 2005-12-22 | | 公開日 | 2006-11-07 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Role of invariant Thr80 in human immunodeficiency virus type 1 protease structure, function, and viral infectivity.
J.Virol., 80, 2006
|
|
2FGU
 
 | | X-ray crystal structure of HIV-1 Protease T80S variant in complex with the inhibitor saquinavir used to explore the role of invariant Thr80 in HIV-1 protease structure, function, and viral infectivity. | | 分子名称: | (2S)-2-amino-3-phenylpropane-1,1-diol, 2-METHYL-DECAHYDRO-ISOQUINOLINE-3-CARBOXYLIC ACID, ASPARAGINE, ... | | 著者 | Foulkes, J.E, Prabu-Jeyabalan, M, Cooper, D, Schiffer, C.A. | | 登録日 | 2005-12-22 | | 公開日 | 2006-11-07 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Role of invariant Thr80 in human immunodeficiency virus type 1 protease structure, function, and viral infectivity.
J.Virol., 80, 2006
|
|
2F3K
 
 | | Substrate envelope and drug resistance: crystal structure of r01 in complex with wild-type hiv-1 protease | | 分子名称: | (3S,4AS,8AS)-N-(TERT-BUTYL)-2-[(3S)-3-({3-(METHYLSULFONYL)-N-[(PYRIDIN-3-YLOXY)ACETYL]-L-VALYL}AMINO)-2-OXO-4-PHENYLBUTYL]DECAHYDROISOQUINOLINE-3-CARBOXAMIDE, PHOSPHATE ION, Protease | | 著者 | Prabu-Jeyabalan, M, King, N.M, Nalivaika, E.A, Heilek-Snyder, G, Cammack, N, Schiffer, C.A. | | 登録日 | 2005-11-21 | | 公開日 | 2006-11-21 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.599 Å) | | 主引用文献 | Substrate envelope and drug resistance: crystal structure of RO1 in complex with wild-type human immunodeficiency virus type 1 protease.
Antimicrob.Agents Chemother., 50, 2006
|
|
4IOU
 
 | |
6VDN
 
 | |
6VDL
 
 | |
6VDM
 
 | |
6VDO
 
 | |
6UE3
 
 | |
6W6Q
 
 | | WT HTLV-1 Protease in Complex with Darunavir (DRV) | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, HTLV-1 Protease | | 著者 | Lockbaum, G.J, Henes, M, Kosovrasti, K, Nalivaika, E.A, Ali, A, KurtYilmaz, N, Schiffer, C.A. | | 登録日 | 2020-03-17 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | To Be Determined
To Be Published
|
|
6W6T
 
 | | WT HIV-1 Protease in Complex with Phosphonated UMass6 (PU6) | | 分子名称: | Protease, SULFATE ION, diethyl [(4-{(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-ethylbutyl)amino}-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxybutyl}phenoxy)methyl]phosphonate | | 著者 | Lockbaum, G.J, Henes, M, Kosovrasti, K, Nalivaika, E.A, Ali, A, KurtYilmaz, N, Schiffer, C.A. | | 登録日 | 2020-03-17 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | To Be Determined
To Be Published
|
|
6W6R
 
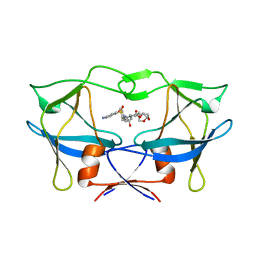 | | WT HTLV-1 Protease in Complex with UMass6 (UM6) | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl](2-ethylbutyl)amino}-1-benzyl-2-hydroxypropyl]carbamate, HTLV-1 Protease | | 著者 | Lockbaum, G.J, Henes, M, Kosovrasti, K, Nalivaika, E.A, Ali, A, KurtYilmaz, N, Schiffer, C.A. | | 登録日 | 2020-03-17 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | To Be Determined
To Be Published
|
|
6W6S
 
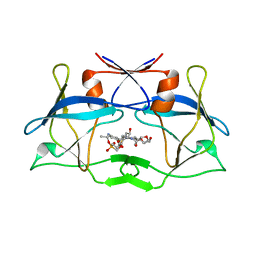 | | WT HTLV-1 Protease in Complex with Phosphonated UMass6 (PU6) | | 分子名称: | HTLV-1 Protease, diethyl [(4-{(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-ethylbutyl)amino}-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxybutyl}phenoxy)methyl]phosphonate | | 著者 | Lockbaum, G.J, Henes, M, Kosovrasti, K, Nalivaika, E.A, Ali, A, KurtYilmaz, N, Schiffer, C.A. | | 登録日 | 2020-03-17 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | To Be Determined
To Be Published
|
|
6WEX
 
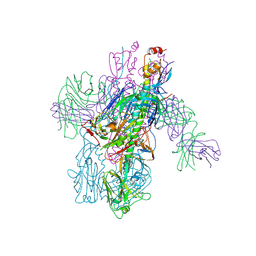 | | Crystal Structure of Broadly Neutralizing Antibody 3I14-D93N Mutant Bound to the Influenza A H6 Hemagglutinin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, Hemagglutinin HA2 chain, ... | | 著者 | Harshbarger, W.D, Lockbaum, G.J, Deming, D.T, Attatippaholkun, N, Schiffer, C.A, Marasco, W.A. | | 登録日 | 2020-04-03 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.49 Å) | | 主引用文献 | Unique structural solution from a V H 3-30 antibody targeting the hemagglutinin stem of influenza A viruses.
Nat Commun, 12, 2021
|
|
3MXD
 
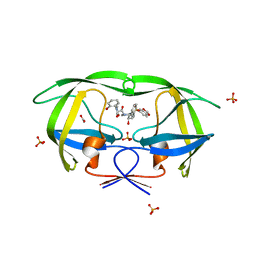 | | Crystal structure of HIV-1 protease inhibitor KC53 in complex with wild-type protease | | 分子名称: | (5S)-N-{(1S,2R)-3-[(1,3-benzodioxol-5-ylsulfonyl)(2-methylpropyl)amino]-1-benzyl-2-hydroxypropyl}-3-(2-hydroxyphenyl)-2 -oxo-1,3-oxazolidine-5-carboxamide, ACETATE ION, HIV-1 protease, ... | | 著者 | Nalam, M.N.L, Schiffer, C.A. | | 登録日 | 2010-05-07 | | 公開日 | 2010-11-10 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure-Based Design, Synthesis, and Structure-Activity Relationship Studies of HIV-1 Protease Inhibitors Incorporating Phenyloxazolidinones.
J.Med.Chem., 53, 2010
|
|
3MXE
 
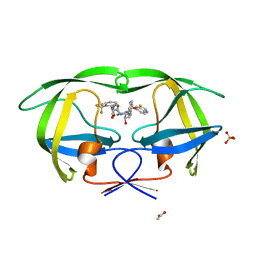 | | Crystal structure of HIV-1 protease inhibitor, KC32 complexed with wild-type protease | | 分子名称: | (5S)-N-{(1S,2R)-3-[(1,3-benzothiazol-6-ylsulfonyl)(2-methylpropyl)amino]-1-benzyl-2-hydroxypropyl}-2-oxo-3-[2-(trifluoromethyl)phenyl]-1,3-oxazolidine-5-carboxamide, ACETATE ION, HIV-1 protease, ... | | 著者 | Nalam, M.N.L, Schiffer, C.A. | | 登録日 | 2010-05-07 | | 公開日 | 2010-11-10 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure-Based Design, Synthesis, and Structure-Activity Relationship Studies of HIV-1 Protease Inhibitors Incorporating Phenyloxazolidinones.
J.Med.Chem., 53, 2010
|
|
4LCE
 
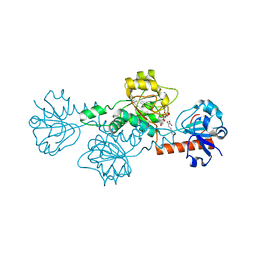 | | CtBP1 in complex with substrate MTOB | | 分子名称: | 4-(METHYLSULFANYL)-2-OXOBUTANOIC ACID, C-terminal-binding protein 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Hilbert, B.J, Schiffer, C.A, Royer Jr, W.E. | | 登録日 | 2013-06-21 | | 公開日 | 2014-03-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Crystal structures of human CtBP in complex with substrate MTOB reveal active site features useful for inhibitor design.
Febs Lett., 588, 2014
|
|
3V4J
 
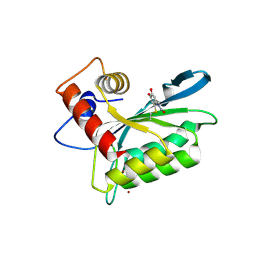 | | First-In-Class Small Molecule Inhibitors of the Single-strand DNA Cytosine Deaminase APOBEC3G | | 分子名称: | 4-[methyl(nitroso)amino]benzene-1,2-diol, DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | 著者 | Shandilya, S.M.D, Ali, A, Schiffer, C.A. | | 登録日 | 2011-12-15 | | 公開日 | 2012-02-01 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | First-In-Class Small Molecule Inhibitors of the Single-Strand DNA Cytosine Deaminase APOBEC3G.
Acs Chem.Biol., 7, 2012
|
|
3V4K
 
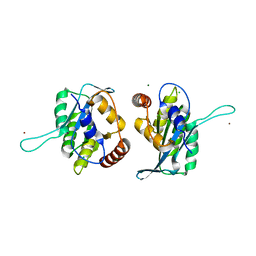 | | First-In-Class Small Molecule Inhibitors of the Single-strand DNA Cytosine Deaminase APOBEC3G | | 分子名称: | CHLORIDE ION, DNA dC->dU-editing enzyme APOBEC-3G, MAGNESIUM ION, ... | | 著者 | Shandilya, S.M.D, Ali, A, Schiffer, C.A. | | 登録日 | 2011-12-15 | | 公開日 | 2012-01-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | First-In-Class Small Molecule Inhibitors of the Single-Strand DNA Cytosine Deaminase APOBEC3G.
Acs Chem.Biol., 7, 2012
|
|
3OXW
 
 | | Crystal Structure of HIV-1 I50V, A71V Protease in Complex with the Protease Inhibitor Darunavir | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, 1,2-ETHANEDIOL, ACETATE ION, ... | | 著者 | Mittal, S, Bandaranayake, R.M, Schiffer, C.A. | | 登録日 | 2010-09-22 | | 公開日 | 2011-09-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural and thermodynamic basis of amprenavir/darunavir and atazanavir resistance in HIV-1 protease with mutations at residue 50.
J.Virol., 87, 2013
|
|
3RC4
 
 | |
