6AIB
 
 | | Crystal structures of the N-terminal RecA-like domain 1 of Staphylococcus aureus DEAD-box Cold shock RNA helicase CshA | | 分子名称: | DEAD-box ATP-dependent RNA helicase CshA | | 著者 | Chengliang, W, Tian, T, Xiaobao, C, Xuan, Z, Jianye, Z. | | 登録日 | 2018-08-22 | | 公開日 | 2018-11-21 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structures of the N-terminal domain of the Staphylococcus aureus DEAD-box RNA helicase CshA and its complex with AMP
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
7FC0
 
 | | Reconstitution of MbnABC complex from Rugamonas rubra ATCC-43154 (GroupIII) | | 分子名称: | FE (III) ION, Methanobactin biosynthesis cassette protein MbnB, Methanobactin biosynthesis cassette protein MbnC, ... | | 著者 | Chao, D, Zhaolin, L, Shoujie, L, Li, Z, Dan, Z, Ying, J, Wei, C. | | 登録日 | 2021-07-13 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.643 Å) | | 主引用文献 | Crystal structure and catalytic mechanism of the MbnBC holoenzyme required for methanobactin biosynthesis.
Cell Res., 32, 2022
|
|
4LMA
 
 | |
4LMB
 
 | |
4AW5
 
 | | Complex of the EphB4 kinase domain with an oxindole inhibitor | | 分子名称: | (3Z)-5-[(1-ethylpiperidin-4-yl)amino]-3-[(5-methoxy-1H-benzimidazol-2-yl)(phenyl)methylidene]-1,3-dihydro-2H-indol-2-one, EPHRIN TYPE-B RECEPTOR 4 | | 著者 | Till, J.H, Stout, T.J. | | 登録日 | 2012-05-31 | | 公開日 | 2012-08-01 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | The Design, Synthesis, and Biological Evaluation of Potent Receptor Tyrosine Kinase Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6INH
 
 | | A glycosyltransferase with UDP and the substrate | | 分子名称: | 1-O-[(8alpha,9beta,10alpha,13alpha)-13-(beta-D-glucopyranosyloxy)-18-oxokaur-16-en-18-yl]-beta-D-glucopyranose, GLYCEROL, UDP-glycosyltransferase 76G1, ... | | 著者 | Zhu, X. | | 登録日 | 2018-10-25 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Hydrophobic recognition allows the glycosyltransferase UGT76G1 to catalyze its substrate in two orientations.
Nat Commun, 10, 2019
|
|
6INI
 
 | | a glycosyltransferase complex with UDP and the product | | 分子名称: | (8alpha,9beta,10alpha,13alpha)-13-{[beta-D-glucopyranosyl-(1->2)-[beta-D-glucopyranosyl-(1->3)]-beta-D-glucopyranosyl]oxy}kaur-16-en-18-oic acid, 1-O-[(8alpha,9beta,10alpha,13alpha)-13-(beta-D-glucopyranosyloxy)-18-oxokaur-16-en-18-yl]-beta-D-glucopyranose, GLYCEROL, ... | | 著者 | Zhu, X, Yang, T, Naismith, J.H. | | 登録日 | 2018-10-25 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Hydrophobic recognition allows the glycosyltransferase UGT76G1 to catalyze its substrate in two orientations.
Nat Commun, 10, 2019
|
|
9BUP
 
 | |
3LFM
 
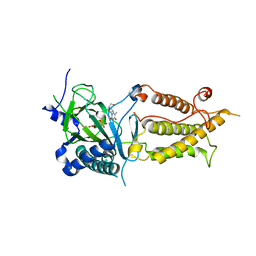 | |
9BUT
 
 | |
9BUS
 
 | |
9BUU
 
 | |
9BUQ
 
 | |
6ING
 
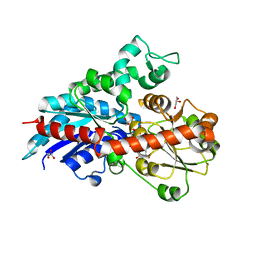 | |
6INF
 
 | | a glycosyltransferase complex with UDP | | 分子名称: | UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE | | 著者 | Zhu, X, Yang, T, Naismith, J.H. | | 登録日 | 2018-10-25 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Hydrophobic recognition allows the glycosyltransferase UGT76G1 to catalyze its substrate in two orientations.
Nat Commun, 10, 2019
|
|
3RN4
 
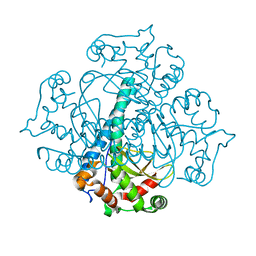 | | Crystal structure of iron-substituted Sod2 from Saccharomyces cerevisiae | | 分子名称: | FE (III) ION, Superoxide dismutase [Mn], mitochondrial | | 著者 | Kang, Y, He, Y.-X, Cheng, W, Zhou, C.-Z, Li, W.-F. | | 登録日 | 2011-04-21 | | 公開日 | 2011-11-23 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Structures of native and Fe-substituted SOD2 from Saccharomyces cerevisiae
Acta Crystallogr.,Sect.F, 67, 2011
|
|
6LT0
 
 | | cryo-EM structure of C9ORF72-SMCR8-WDR41 | | 分子名称: | Guanine nucleotide exchange C9orf72, Guanine nucleotide exchange protein SMCR8, WD repeat-containing protein 41 | | 著者 | Tang, D, Sheng, J, Xu, L, Zhan, X, Yan, C, Qi, S. | | 登録日 | 2020-01-21 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structure of C9ORF72-SMCR8-WDR41 reveals the role as a GAP for Rab8a and Rab11a.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7XWX
 
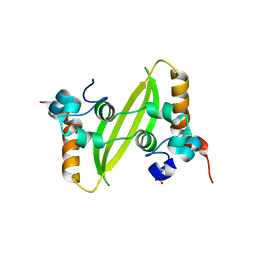 | | Crystal structure of SARS-CoV-2 N-CTD | | 分子名称: | Nucleoprotein, PHOSPHATE ION | | 著者 | Luan, X.D, Li, X.M, Li, Y.F. | | 登録日 | 2022-05-27 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Antiviral drug design based on structural insights into the N-terminal domain and C-terminal domain of the SARS-CoV-2 nucleocapsid protein.
Sci Bull (Beijing), 67, 2022
|
|
7XX1
 
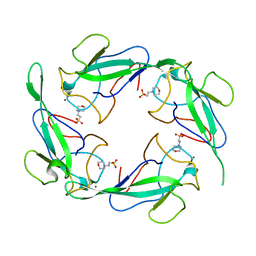 | | Crystal structure of SARS-CoV-2 N-NTD | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nucleoprotein, ZINC ION | | 著者 | Luan, X.D, Li, X.M, Li, Y.F. | | 登録日 | 2022-05-27 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Antiviral drug design based on structural insights into the N-terminal domain and C-terminal domain of the SARS-CoV-2 nucleocapsid protein.
Sci Bull (Beijing), 67, 2022
|
|
7XWZ
 
 | | Crystal structure of SARS-CoV-2 N-NTD and dsRNA complex | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Nucleoprotein, ... | | 著者 | Luan, X.D, Li, X.M, Li, Y.F. | | 登録日 | 2022-05-27 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Antiviral drug design based on structural insights into the N-terminal domain and C-terminal domain of the SARS-CoV-2 nucleocapsid protein.
Sci Bull (Beijing), 67, 2022
|
|
6IZK
 
 | | Structural characterization of mutated NreA protein in nitrate binding site from Staphylococcus aureus | | 分子名称: | CHLORIDE ION, IMIDAZOLE, L(+)-TARTARIC ACID, ... | | 著者 | Sangare, L, Chen, W, Wang, C, Chen, X, Wu, M, Zhang, X, Zang, J. | | 登録日 | 2018-12-19 | | 公開日 | 2020-01-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Structural characterization of mutated NreA protein in nitrate binding site from Staphylococcus aureus
To Be Published
|
|
4OI6
 
 | | Crystal structure analysis of nickel-bound form SCO4226 from Streptomyces coelicolor A3(2) | | 分子名称: | CITRIC ACID, NICKEL (II) ION, Nickel responsive protein | | 著者 | Lu, M, Jiang, Y.L, Wang, S, Cheng, W, Zhang, R.G, Virolle, M.J, Chen, Y, Zhou, C.Z. | | 登録日 | 2014-01-18 | | 公開日 | 2014-09-10 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Streptomyces coelicolor SCO4226 Is a Nickel Binding Protein.
Plos One, 9, 2014
|
|
4OI3
 
 | | Crystal structure analysis of SCO4226 from Streptomyces coelicolor A3(2) | | 分子名称: | Nickel responsive protein | | 著者 | Lu, M, Jiang, Y.L, Wang, S, Cheng, W, Zhang, R.G, Virolle, M.J, Chen, Y, Zhou, C.Z. | | 登録日 | 2014-01-18 | | 公開日 | 2014-09-17 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Streptomyces coelicolor SCO4226 Is a Nickel Binding Protein.
Plos One, 9, 2014
|
|
7DZ9
 
 | | MbnABC complex | | 分子名称: | FE (III) ION, MbnA, MbnB, ... | | 著者 | Chao, D, Dan, Z, Yijun, G, Wei, C. | | 登録日 | 2021-01-25 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure and catalytic mechanism of the MbnBC holoenzyme required for methanobactin biosynthesis.
Cell Res., 32, 2022
|
|
7ERY
 
 | | apo form of the glycosyltransferase | | 分子名称: | Glycosyltransferase | | 著者 | Zhu, X. | | 登録日 | 2021-05-08 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides.
Nat Commun, 12, 2021
|
|
