8DGY
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 16d (high resolution) | | 分子名称: | 3C-like proteinase, [(1~{R},2~{R})-2-(cyclohexylmethyl)cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, [(1~{R},2~{R})-2-(cyclohexylmethyl)cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | 著者 | Lovell, S, Liu, L, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | 登録日 | 2022-06-24 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8D9C
 
 | | Crystal Structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with fluorinated inhibitor 10 | | 分子名称: | 2,3,4,5,6-pentafluoro-N-hydroxybenzamide, Hdac6 protein, POTASSIUM ION, ... | | 著者 | Watson, P.R, Christianson, D.W. | | 登録日 | 2022-06-09 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Aromatic Ring Fluorination Patterns Modulate Inhibitory Potency of Fluorophenylhydroxamates Complexed with Histone Deacetylase 6.
Biochemistry, 61, 2022
|
|
8D99
 
 | | Crystal Structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with fluorinated inhibitor 7 | | 分子名称: | 2,3,6-trifluoro-N-hydroxybenzamide, Hdac6 protein, POTASSIUM ION, ... | | 著者 | Watson, P.R, Christianson, D.W. | | 登録日 | 2022-06-09 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Aromatic Ring Fluorination Patterns Modulate Inhibitory Potency of Fluorophenylhydroxamates Complexed with Histone Deacetylase 6.
Biochemistry, 61, 2022
|
|
8D9B
 
 | | Crystal Structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with fluorinated inhibitor 9 | | 分子名称: | 2,3,5,6-tetrafluoro-N-hydroxybenzamide, Hdac6 protein, POTASSIUM ION, ... | | 著者 | Watson, P.R, Christianson, D.W. | | 登録日 | 2022-06-09 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Aromatic Ring Fluorination Patterns Modulate Inhibitory Potency of Fluorophenylhydroxamates Complexed with Histone Deacetylase 6.
Biochemistry, 61, 2022
|
|
8D9A
 
 | | Crystal Structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with fluorinated inhibitor 8 | | 分子名称: | 2,3,5-trifluoro-N-hydroxybenzamide, Hdac6 protein, POTASSIUM ION, ... | | 著者 | Watson, P.R, Christianson, D.W. | | 登録日 | 2022-06-09 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Aromatic Ring Fluorination Patterns Modulate Inhibitory Potency of Fluorophenylhydroxamates Complexed with Histone Deacetylase 6.
Biochemistry, 61, 2022
|
|
8D98
 
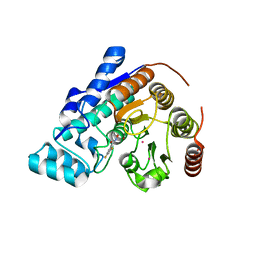 | |
7JYY
 
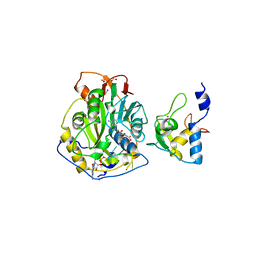 | | Crystal Structure of SARS-CoV-2 Nsp16/10 Heterodimer in Complex with (m7GpppA)pUpUpApApA (Cap-0) and S-Adenosylmethionine (SAM). | | 分子名称: | 2'-O-methyltransferase, CHLORIDE ION, FORMIC ACID, ... | | 著者 | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-09-01 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Mn 2+ coordinates Cap-0-RNA to align substrates for efficient 2'- O -methyl transfer by SARS-CoV-2 nsp16.
Sci.Signal., 14, 2021
|
|
7LRD
 
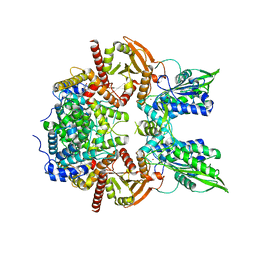 | | Cryo-EM of the SLFN12-PDE3A complex: Consensus subset model | | 分子名称: | (4~{R})-3-[4-(diethylamino)-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]-4-methyl-4,5-dihydro-1~{H}-pyridazin-6-one, MAGNESIUM ION, MANGANESE (II) ION, ... | | 著者 | Fuller, J.R, Garvie, C.W, Lemke, C.T. | | 登録日 | 2021-02-16 | | 公開日 | 2021-06-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.22 Å) | | 主引用文献 | Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase.
Nat Commun, 12, 2021
|
|
7LRE
 
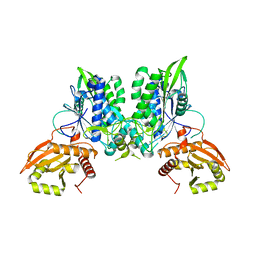 | |
7LRC
 
 | | Cryo-EM of the SLFN12-PDE3A complex: PDE3A body refinement | | 分子名称: | (4~{R})-3-[4-(diethylamino)-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]-4-methyl-4,5-dihydro-1~{H}-pyridazin-6-one, MAGNESIUM ION, MANGANESE (II) ION, ... | | 著者 | Fuller, J.R, Garvie, C.W, Lemke, C.T. | | 登録日 | 2021-02-16 | | 公開日 | 2021-06-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (2.97 Å) | | 主引用文献 | Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase.
Nat Commun, 12, 2021
|
|
7JWL
 
 | | Crystal Structure of Pseudomonas aeruginosa Penicillin Binding Protein 3 (PAE-PBP3) bound to ETX0462 | | 分子名称: | CHLORIDE ION, ETX0462 (Bound form), Peptidoglycan D,D-transpeptidase FtsI | | 著者 | Mayclin, S.J, Abendroth, J, Horanyi, P.S, Sylvester, M, Wu, X, Shapiro, A, Moussa, S, Durand-Reville, T.F. | | 登録日 | 2020-08-25 | | 公開日 | 2021-05-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Rational design of a new antibiotic class for drug-resistant infections.
Nature, 597, 2021
|
|
7JOM
 
 | |
8CYZ
 
 | | Crystal structure of SARS-CoV-2 Mpro with compound C4 | | 分子名称: | 3C-like proteinase, N-[(4-chlorothiophen-2-yl)methyl]-2-(isoquinolin-4-yl)-N-[4-(methylsulfanyl)phenyl]acetamide | | 著者 | Worrall, L.J, Lee, J, Strynadka, N.C.J. | | 登録日 | 2022-05-24 | | 公開日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
8CZ7
 
 | | Crystal structure of SARS-CoV-2 Mpro with compound C2 | | 分子名称: | 3C-like proteinase, N-[(4-chlorothiophen-2-yl)methyl]-2-(isoquinolin-4-yl)-N-(4-methoxyphenyl)acetamide | | 著者 | Worrall, L.J, Lee, J, Strynadka, N.C.J. | | 登録日 | 2022-05-24 | | 公開日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
7C0Q
 
 | | a Legionella pneumophila effector Lpg2505 | | 分子名称: | GLYCEROL, effector Lpg2505 | | 著者 | Chen, T.T, Han, A.D, Luo, Z.Q, McCloskey, A, Perri, K. | | 登録日 | 2020-05-01 | | 公開日 | 2021-02-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The metaeffector MesI regulates the activity of the Legionella effector SidI through direct protein-protein interactions.
Microbes Infect., 23, 2021
|
|
8CYU
 
 | | Crystal structure of SARS-CoV-2 Mpro with compound C5 | | 分子名称: | 3C-like proteinase, N-[(4-chlorothiophen-2-yl)methyl]-N-[4-(dimethylamino)phenyl]-2-(isoquinolin-4-yl)acetamide | | 著者 | Worrall, L.J, Lee, J, Strynadka, N.C.J. | | 登録日 | 2022-05-24 | | 公開日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
8CZ4
 
 | | Crystal structure of SARS-CoV-2 Mpro with compound C3 | | 分子名称: | 3C-like proteinase, N-(4-tert-butylphenyl)-N-[(4-chlorothiophen-2-yl)methyl]-2-(isoquinolin-4-yl)acetamide | | 著者 | Worrall, L.J, Lee, J, Strynadka, N.C.J. | | 登録日 | 2022-05-24 | | 公開日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
7KQ1
 
 | | PCNA bound to truncated peptide mimetic | | 分子名称: | LYS-ARG-ARG-GLN-THR-SER-MET-THR-ASP-PHE-TYR-HIS-SER-LYS-ARG, Proliferating cell nuclear antigen | | 著者 | Vandborg, B.A, Bruning, J.B. | | 登録日 | 2020-11-13 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Unlocking the PIP-box: A peptide library reveals interactions that drive high-affinity binding to human PCNA.
J.Biol.Chem., 296, 2021
|
|
7KQ0
 
 | | PCNA bound to peptide mimetic | | 分子名称: | LYS-ARG-ARG-GLN-THR-SER-MET-THR-ASP-TYR-TYR-HIS-SER-LYS-ARG, Proliferating cell nuclear antigen | | 著者 | Vandborg, B.A, Bruning, J.B. | | 登録日 | 2020-11-13 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Unlocking the PIP-box: A peptide library reveals interactions that drive high-affinity binding to human PCNA.
J.Biol.Chem., 296, 2021
|
|
7LAD
 
 | | Clobetasol propionate bound to CYP3A5 | | 分子名称: | Clobetasol propionate, Cytochrome P450 3A5, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Buchman, C.D, Miller, D, Wang, J, Jayaraman, S, Chen, T. | | 登録日 | 2021-01-06 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Unraveling the Structural Basis of Selective Inhibition of Human Cytochrome P450 3A5.
J.Am.Chem.Soc., 143, 2021
|
|
8E2M
 
 | | Bruton's tyrosine kinase (BTK) with compound 13 | | 分子名称: | (5P)-5-[4-methyl-6-(2-methylpropyl)pyridin-3-yl]-4-oxo-N-[(1R,2S)-2-propanamidocyclopentyl]-4,5-dihydro-3H-1-thia-3,5,8-triazaacenaphthylene-2-carboxamide, TRIETHYLENE GLYCOL, Tyrosine-protein kinase BTK | | 著者 | Alexander, R, Milligan, C.M. | | 登録日 | 2022-08-15 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.904 Å) | | 主引用文献 | Discovery of JNJ-64264681: A Potent and Selective Covalent Inhibitor of Bruton's Tyrosine Kinase.
J.Med.Chem., 65, 2022
|
|
8E5U
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 9 | | 分子名称: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-(6-{2-nitro-4-[(1R,5S)-3-oxa-8-azabicyclo[3.2.1]octan-8-yl]anilino}hexyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Karade, S.S, Mariuzza, R.A. | | 登録日 | 2022-08-22 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8E3J
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 4 | | 分子名称: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-{6-[2-nitro-4-(pyrimidin-2-yl)anilino]hexyl}piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, ... | | 著者 | Karade, S.S, Mariuzza, R.A. | | 登録日 | 2022-08-17 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8E3P
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 5 | | 分子名称: | (2S,3S,4S,5R)-2-(hydroxymethyl)-1-{6-[3-nitro-5-(pyridin-4-yl)anilino]hexyl}piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, ... | | 著者 | Karade, S.S, Mariuzza, R.A. | | 登録日 | 2022-08-17 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8E4Z
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 8 | | 分子名称: | (2R,3R,4R,5S)-1-(6-{[(4P)-4-(5-cyclobutyl-1,2,4-oxadiazol-3-yl)-2-nitrophenyl]amino}hexyl)-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Karade, S.S, Mariuzza, R.A. | | 登録日 | 2022-08-19 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
