7RM2
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with Mcule-CSR-494190-S1 | | 分子名称: | 3C-like proteinase, 6-[4-(3,5-dichloro-4-methylphenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-26 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RME
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-52 | | 分子名称: | 3C-like proteinase, 6-{4-[4-chloro-3-(trifluoromethyl)phenyl]piperazine-1-carbonyl}pyrimidine-2,4(1H,3H)-dione | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-27 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RMT
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-70 | | 分子名称: | 2-chloro-4-[4-(2,6-dioxo-1,2,5,6-tetrahydropyrimidine-4-carbonyl)piperazin-1-yl]benzaldehyde, 3C-like proteinase | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-28 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RNH
 
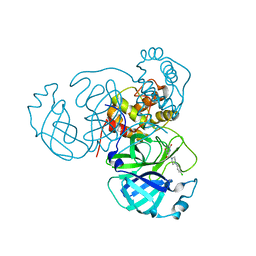 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-45 | | 分子名称: | 3C-like proteinase, 6-[4-(4-chlorophenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-29 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RMB
 
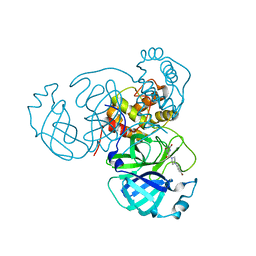 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-78 | | 分子名称: | 3C-like proteinase, 6-[4-(4-bromo-3-chlorophenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-27 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RN4
 
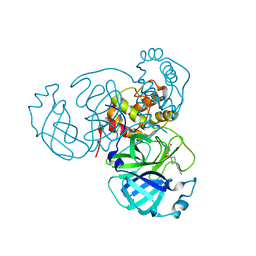 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-69 | | 分子名称: | 3C-like proteinase, 6-[4-(3,4-dichlorophenyl)piperidine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-29 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
1G3G
 
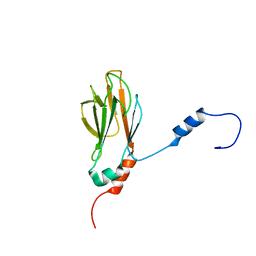 | | NMR STRUCTURE OF THE FHA1 DOMAIN OF YEAST RAD53 | | 分子名称: | PROTEIN KINASE SPK1 | | 著者 | Yuan, C, Liao, H, Su, M, Yongkiettrakul, S, Byeon, I.-J.L, Tsai, M.-D. | | 登録日 | 2000-10-24 | | 公開日 | 2001-01-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the FHA1 domain of yeast Rad53 and identification of binding sites for both FHA1 and its target protein Rad9
J.Mol.Biol., 304, 2000
|
|
7RMZ
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-63 | | 分子名称: | 3C-like proteinase, 6-{4-[3-chloro-4-(trifluoromethyl)phenyl]piperazine-1-carbonyl}pyrimidine-2,4(1H,3H)-dione | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-28 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
5VYG
 
 | | Crystal structure of hFA9 EGF repeat with O-glucose trisaccharide | | 分子名称: | CALCIUM ION, Coagulation factor IX, alpha-D-xylopyranose-(1-3)-alpha-D-xylopyranose-(1-3)-beta-D-glucopyranose | | 著者 | Yu, H.J, Li, H.L. | | 登録日 | 2017-05-25 | | 公開日 | 2017-08-09 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | O-Glycosylation modulates the stability of epidermal growth factor-like repeats and thereby regulates Notch trafficking
J. Biol. Chem., 292, 2017
|
|
8Y7L
 
 | |
1QO3
 
 | |
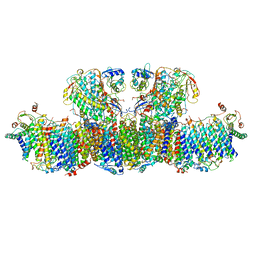
6U8Y
 
 | |
7N8C
 
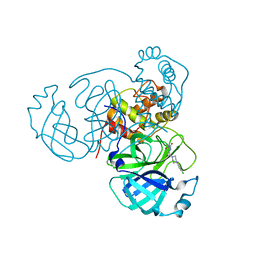 | | Joint X-ray/neutron structure of SARS-CoV-2 main protease (Mpro) in complex with Mcule5948770040 | | 分子名称: | 3C-like proteinase, 6-[4-(3,4-dichlorophenyl)piperazin-1-yl]carbonyl-1~{H}-pyrimidine-2,4-dione | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-06-14 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | NEUTRON DIFFRACTION (2.2 Å), X-RAY DIFFRACTION | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
3LKZ
 
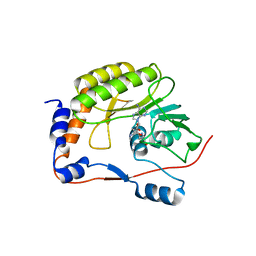 | |
5GJ9
 
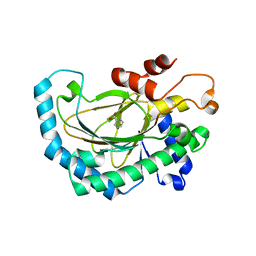 | | Crystal structure of Arabidopsis thaliana ACO2 in complex with POA | | 分子名称: | 1-aminocyclopropane-1-carboxylate oxidase 2, PYRAZINE-2-CARBOXYLIC ACID, ZINC ION | | 著者 | Sun, X.Z, Li, Y.X, He, W.R, Ji, C.G, Xia, P.X, Wang, Y.C, Du, S, Li, H.J, Raikhel, N, Xiao, J.Y, Guo, H.W. | | 登録日 | 2016-06-28 | | 公開日 | 2017-05-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Pyrazinamide and derivatives block ethylene biosynthesis by inhibiting ACC oxidase.
Nat Commun, 8, 2017
|
|
6SZE
 
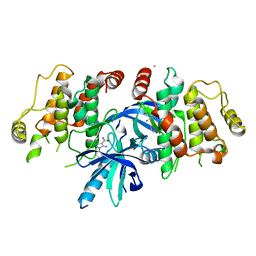 | |
6SZJ
 
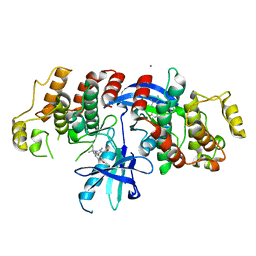 | |
3PDV
 
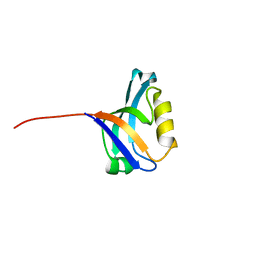 | |
5GJA
 
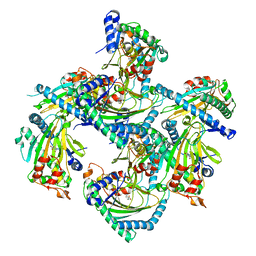 | | Crystal structure of Arabidopsis thaliana ACO2 in complex with 2-PA | | 分子名称: | 1-aminocyclopropane-1-carboxylate oxidase 2, PYRIDINE-2-CARBOXYLIC ACID, ZINC ION | | 著者 | Sun, X.Z, Li, Y.X, He, W.R, Ji, C.G, Xia, P.X, Wang, Y.C, Du, S, Li, H.J, Raikhel, N, Xiao, J.Y, Guo, H.W. | | 登録日 | 2016-06-28 | | 公開日 | 2017-05-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Pyrazinamide and derivatives block ethylene biosynthesis by inhibiting ACC oxidase.
Nat Commun, 8, 2017
|
|
6D7J
 
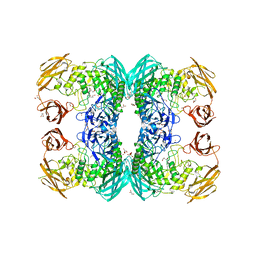 | |
4N0S
 
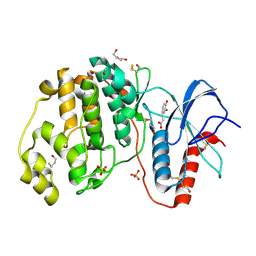 | | Complex of ERK2 with caffeic acid | | 分子名称: | CAFFEIC ACID, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | 著者 | Kurinov, I, Malakhova, M. | | 登録日 | 2013-10-02 | | 公開日 | 2014-08-27 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.7992 Å) | | 主引用文献 | Caffeic Acid Directly Targets ERK1/2 to Attenuate Solar UV-Induced Skin Carcinogenesis.
Cancer Prev Res (Phila), 7, 2014
|
|
1MLU
 
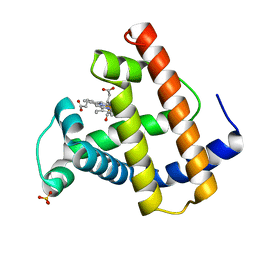 | |
6LNA
 
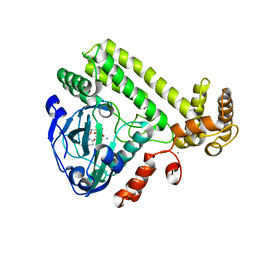 | | YdiU complex with AMPNPP and Mn2+ | | 分子名称: | CALCIUM ION, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Li, B, Yang, Y, Ma, Y. | | 登録日 | 2019-12-28 | | 公開日 | 2020-12-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.701 Å) | | 主引用文献 | The YdiU Domain Modulates Bacterial Stress Signaling through Mn 2+ -Dependent UMPylation.
Cell Rep, 32, 2020
|
|
4YFI
 
 | | TNNI3K complexed with inhibitor 1 | | 分子名称: | N-methyl-3-(9H-purin-6-ylamino)benzenesulfonamide, Serine/threonine-protein kinase TNNI3K | | 著者 | Shewchuk, L.M, Wang, L, Lawhorn, B.G. | | 登録日 | 2015-02-25 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Identification of Purines and 7-Deazapurines as Potent and Selective Type I Inhibitors of Troponin I-Interacting Kinase (TNNI3K).
J.Med.Chem., 58, 2015
|
|
4YFF
 
 | | TNNI3K complexed with inhibitor 2 | | 分子名称: | 3-[(5-bromo-7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]-N-methyl-4-(morpholin-4-yl)benzenesulfonamide, Serine/threonine-protein kinase TNNI3K | | 著者 | Shewchuk, L.M, Wang, L, Lawhorn, B.G. | | 登録日 | 2015-02-25 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.07 Å) | | 主引用文献 | Identification of Purines and 7-Deazapurines as Potent and Selective Type I Inhibitors of Troponin I-Interacting Kinase (TNNI3K).
J.Med.Chem., 58, 2015
|
|
