8GWN
 
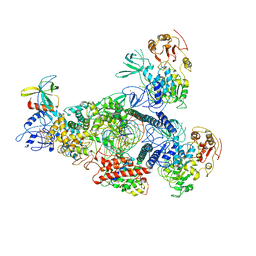 | | A mechanism for SARS-CoV-2 RNA capping and its inhibitor of AT-527 | | 分子名称: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | 著者 | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | 登録日 | 2022-09-17 | | 公開日 | 2022-12-14 | | 最終更新日 | 2023-10-25 | | 実験手法 | ELECTRON MICROSCOPY (3.38 Å) | | 主引用文献 | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
4Y5W
 
 | | Transcription factor-DNA complex | | 分子名称: | DNA (5'-D(P*AP*TP*GP*GP*AP*TP*TP*TP*CP*CP*TP*AP*GP*GP*AP*AP*GP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*CP*TP*TP*CP*CP*TP*AP*GP*GP*AP*AP*AP*TP*CP*CP*AP*T)-3'), Signal transducer and activator of transcription 6 | | 著者 | Li, J, Niu, F, Ouyang, S, Liu, Z. | | 登録日 | 2015-02-12 | | 公開日 | 2016-02-17 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.104 Å) | | 主引用文献 | Structural basis for DNA recognition by STAT6
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8GTK
 
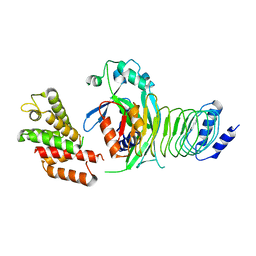 | |
8GTN
 
 | |
8GTJ
 
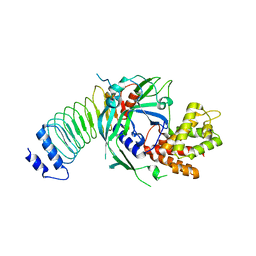 | |
4ZUU
 
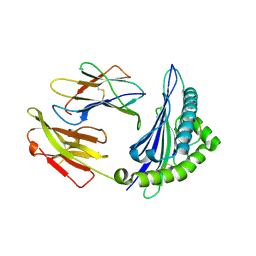 | | Crystal structure of Equine MHC I(Eqca-N*00602) in complexed with equine infectious anaemia virus (EIAV) derived peptide Gag-CF9 | | 分子名称: | Beta-2-microglobulin, CYS-THR-SER-GLU-GLU-MET-ASN-ALA-PHE, Classical MHC class I antigen | | 著者 | Yao, S, Liu, J, Qi, J, Chen, R, Zhang, N, Liu, Y, Xia, C. | | 登録日 | 2015-05-17 | | 公開日 | 2016-04-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Illumination of Equine MHC Class I Molecules Highlights Unconventional Epitope Presentation Manner That Is Evolved in Equine Leukocyte Antigen Alleles
J Immunol., 196, 2016
|
|
4ZUV
 
 | | Crystal structure of Equine MHC I(Eqca-N*00602) in complexed with equine infectious anaemia virus (EIAV) derived peptide Env-RW12 | | 分子名称: | ARG-VAL-GLU-ASP-VAL-THR-ASN-THR-ALA-GLU-TYR-TRP, Beta-2-microglobulin, Classical MHC class I antigen | | 著者 | Yao, S, Liu, J, Qi, J, Chen, R, Zhang, N, Liu, Y, Xia, C. | | 登録日 | 2015-05-17 | | 公開日 | 2016-04-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Illumination of Equine MHC Class I Molecules Highlights Unconventional Epitope Presentation Manner That Is Evolved in Equine Leukocyte Antigen Alleles
J Immunol., 196, 2016
|
|
7JHQ
 
 | | OXA-48 bound by Compound 2.3 | | 分子名称: | 1,2-ETHANEDIOL, Beta-lactamase OXA-48, CHLORIDE ION, ... | | 著者 | Taylor, D.M, Hu, L, Prasad, B.V.V, Palzkill, T. | | 登録日 | 2020-07-21 | | 公開日 | 2021-12-01 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
5FFE
 
 | |
7XPQ
 
 | |
7XPO
 
 | | Crystal Structure of UDP-Glc/GlcNAc 4-Epimerase with NAD/UDP-Glc | | 分子名称: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase, ... | | 著者 | Chen, Y.H, Wang, X.C, Zhang, C.R. | | 登録日 | 2022-05-05 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | A maize epimerase modulates cell wall synthesis and glycosylation during stomatal morphogenesis.
Nat Commun, 14, 2023
|
|
7XPP
 
 | |
5FFD
 
 | |
5FFB
 
 | | CopM in the apo form | | 分子名称: | CopM | | 著者 | Zhao, S, Wang, X, Liu, L. | | 登録日 | 2015-12-18 | | 公開日 | 2016-09-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.702 Å) | | 主引用文献 | Structural basis for copper/silver binding by the Synechocystis metallochaperone CopM.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5YUD
 
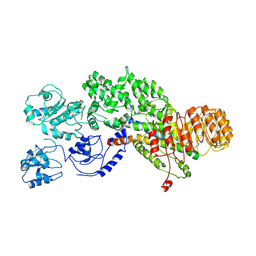 | | Flagellin derivative in complex with the NLR protein NAIP5 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Baculoviral IAP repeat-containing protein 1e, Phase 2 flagellin,Flagellin | | 著者 | Yang, X.R, Yang, F, Wang, W.G, Lin, G.Z. | | 登録日 | 2017-11-21 | | 公開日 | 2018-01-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.28 Å) | | 主引用文献 | Structural basis for specific flagellin recognition by the NLR protein NAIP5.
Cell Res., 28, 2018
|
|
4ZUT
 
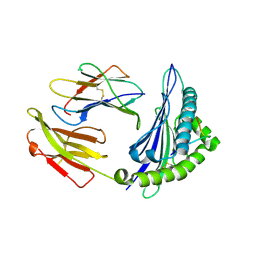 | | Crystal structure of Equine MHC I(Eqca-N*00602) in complexed with equine infectious anaemia virus (EIAV) derived peptide Gag-GW12 | | 分子名称: | Beta-2-microglobulin, Classical MHC class I antigen, GLY-SER-GLN-LYS-LEU-THR-THR-GLY-ASN-CYS-ASN-TRP | | 著者 | Yao, S, Liu, J, Qi, J, Chen, R, Zhang, N, Liu, Y, Xia, C. | | 登録日 | 2015-05-17 | | 公開日 | 2016-04-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural Illumination of Equine MHC Class I Molecules Highlights Unconventional Epitope Presentation Manner That Is Evolved in Equine Leukocyte Antigen Alleles
J Immunol., 196, 2016
|
|
8GWK
 
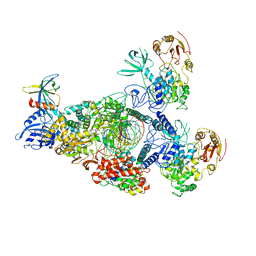 | | SARS-CoV-2 RNA E-RTC complex with RMP-nsp9 and GMPPNP | | 分子名称: | Helicase, MAGNESIUM ION, Non-structural protein 7, ... | | 著者 | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | 登録日 | 2022-09-17 | | 公開日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (2.72 Å) | | 主引用文献 | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8IG7
 
 | |
8IG4
 
 | |
8IG5
 
 | | Crystal structure of SARS main protease in complex with GC376 | | 分子名称: | 3C-like proteinase nsp5, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | 著者 | Lin, C, Zhang, J, Li, J. | | 登録日 | 2023-02-20 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Structural Basis for Coronaviral Main Proteases Inhibition by the 3CLpro Inhibitor GC376.
J.Mol.Biol., 436, 2024
|
|
8IG6
 
 | |
8IG8
 
 | |
8IG9
 
 | |
6QN3
 
 | |
7Y4F
 
 | | bacterial DPP4 | | 分子名称: | Dipeptidyl peptidase IV | | 著者 | Hang, J, Jiang, C, Wang, K, Zhang, Z, Guo, F, Liu, J, Wang, G, Lei, X, Gonzalez, F, Qiao, J. | | 登録日 | 2022-06-14 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.918 Å) | | 主引用文献 | Microbial-host-isozyme analyses reveal microbial DPP4 as a potential antidiabetic target.
Science, 381, 2023
|
|
