7WPH
 
 | | SARS-CoV2 RBD bound to Fab06 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB06 light chain, Fab06 heavy chain, ... | | 著者 | Lin, J.Q, El Sahili, A, Lescar, J. | | 登録日 | 2022-01-23 | | 公開日 | 2022-03-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Engineering SARS-CoV-2 specific cocktail antibodies into a bispecific format improves neutralizing potency and breadth.
Nat Commun, 13, 2022
|
|
7WPV
 
 | |
6BHI
 
 | | Crystal structure of SETDB1 with a modified H3 peptide | | 分子名称: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, UNKNOWN ATOM OR ION | | 著者 | Qin, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2017-10-30 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
6BHD
 
 | | Crystal structure of SETDB1 with a modified H3 peptide | | 分子名称: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, SODIUM ION, ... | | 著者 | Qin, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2017-10-30 | | 公開日 | 2017-12-06 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
6BHH
 
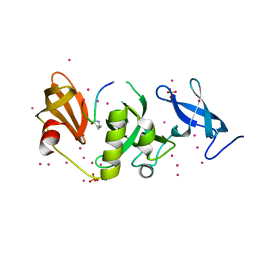 | | Crystal structure of SETDB1 with a modified H3 peptide | | 分子名称: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, SULFATE ION, ... | | 著者 | Qin, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2017-10-30 | | 公開日 | 2017-11-22 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
6BHE
 
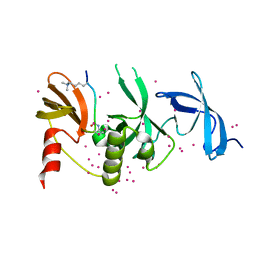 | | Crystal structure of SETDB1 with a modified H3 peptide | | 分子名称: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, UNKNOWN ATOM OR ION | | 著者 | Qin, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2017-10-30 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
7WJM
 
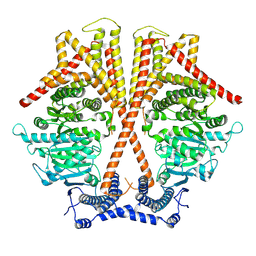 | |
7WJN
 
 | | CryoEM structure of chitin synthase 1 mutant E495A from Phytophthora sojae complexed with UDP-GlcNAc | | 分子名称: | Chitin synthase, MANGANESE (II) ION, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | 著者 | Chen, W, Cao, P, Gong, Y, Yang, Q. | | 登録日 | 2022-01-07 | | 公開日 | 2022-09-28 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis for directional chitin biosynthesis.
Nature, 610, 2022
|
|
7WJO
 
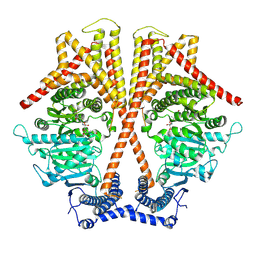 | | CryoEM structure of chitin synthase 1 from Phytophthora sojae complexed with nikkomycin Z | | 分子名称: | (2S)-{[(2S,3S,4S)-2-amino-4-hydroxy-4-(5-hydroxypyridin-2-yl)-3-methylbutanoyl]amino}[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]acetic acid (non-preferred name), Chitin synthase | | 著者 | Chen, W, Cao, P, Gong, Y, Yang, Q. | | 登録日 | 2022-01-07 | | 公開日 | 2022-09-28 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis for directional chitin biosynthesis.
Nature, 610, 2022
|
|
7EN6
 
 | |
7EN5
 
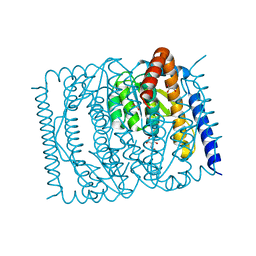 | | The crystal structure of Escherichia coli MurR in complex with N-acetylglucosamine-6-phosphate | | 分子名称: | 2-METHOXYETHANOL, 2-acetamido-2-deoxy-6-O-phosphono-beta-D-glucopyranose, GLYCEROL, ... | | 著者 | Zhang, Y, Chen, W, Ji, Q. | | 登録日 | 2021-04-16 | | 公開日 | 2022-04-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Molecular basis for cell-wall recycling regulation by transcriptional repressor MurR in Escherichia coli.
Nucleic Acids Res., 50, 2022
|
|
7EN7
 
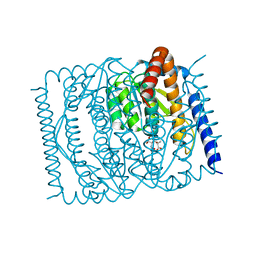 | | The crystal structure of Escherichia coli MurR in complex with N-acetylmuramic-acid-6-phosphate | | 分子名称: | (2R)-2-[(2R,3R,4R,5S,6R)-3-acetamido-2,5-bis(oxidanyl)-6-(phosphonooxymethyl)oxan-4-yl]oxypropanoic acid, HTH-type transcriptional regulator MurR | | 著者 | Zhang, Y, Chen, W, Ji, Q. | | 登録日 | 2021-04-16 | | 公開日 | 2022-04-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | Molecular basis for cell-wall recycling regulation by transcriptional repressor MurR in Escherichia coli.
Nucleic Acids Res., 50, 2022
|
|
7BU9
 
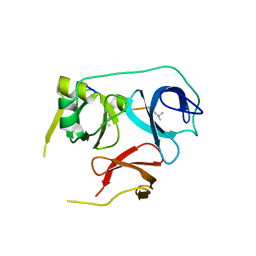 | |
7BQZ
 
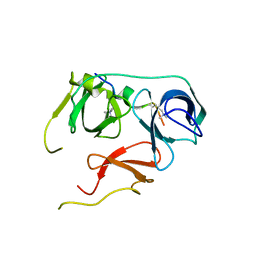 | |
7BTD
 
 | |
7CPC
 
 | |
7CPI
 
 | |
7DCN
 
 | |
7DCM
 
 | | Crystal structure of CITX | | 分子名称: | Probable apo-citrate lyase phosphoribosyl-dephospho-CoA transferase, ZINC ION | | 著者 | Xu, H, Wang, B, Su, X.D. | | 登録日 | 2020-10-26 | | 公開日 | 2021-11-03 | | 最終更新日 | 2023-05-17 | | 実験手法 | X-RAY DIFFRACTION (2.495 Å) | | 主引用文献 | Co-evolution-based prediction of metal-binding sites in proteomes by machine learning.
Nat.Chem.Biol., 19, 2023
|
|
7DNR
 
 | |
7E5C
 
 | | Bacterial prolidase mutant D45W/L225Y/H226L/H343I | | 分子名称: | MANGANESE (II) ION, Xaa-Pro dipeptidase | | 著者 | Yang, J. | | 登録日 | 2021-02-18 | | 公開日 | 2021-03-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Structure-based redesign of the bacterial prolidase active-site pocket for efficient enhancement of methyl-parathion hydrolysis
Catalysis Science And Technology, 11, 2021
|
|
7DWB
 
 | | Human Pannexin1 model | | 分子名称: | Pannexin-1 | | 著者 | Zhang, S.S, Yang, M.J. | | 登録日 | 2021-01-17 | | 公開日 | 2021-05-26 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Structure of the full-length human Pannexin1 channel and insights into its role in pyroptosis.
Cell Discov, 7, 2021
|
|
7DX4
 
 | | The structure of FC08 Fab-hA.CE2-RBD complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Heavy chain of FC08 Fab, ... | | 著者 | Cao, L, Wang, X. | | 登録日 | 2021-01-18 | | 公開日 | 2021-04-21 | | 最終更新日 | 2022-02-23 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | A proof of concept for neutralizing antibody-guided vaccine design against SARS-CoV-2.
Natl Sci Rev, 8, 2021
|
|
7D4G
 
 | |
7BTA
 
 | |
