2HS1
 
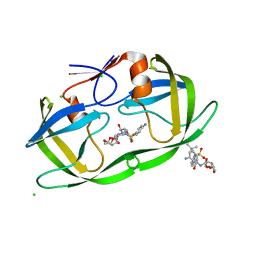 | | Ultra-high resolution X-ray crystal structure of HIV-1 protease V32I mutant with TMC114 (darunavir) inhibitor | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Weber, I.T, Kovalevsky, A.Y. | | 登録日 | 2006-07-20 | | 公開日 | 2006-10-03 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (0.84 Å) | | 主引用文献 | Ultra-high Resolution Crystal Structure of HIV-1 Protease Mutant Reveals Two Binding Sites for Clinical Inhibitor TMC114.
J.Mol.Biol., 363, 2006
|
|
2HUJ
 
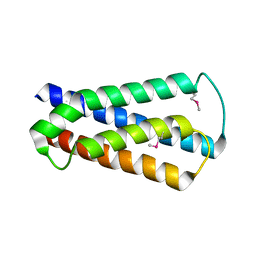 | |
3EO1
 
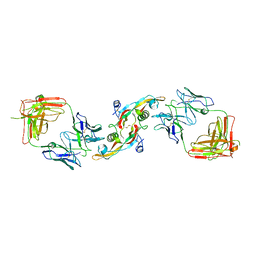 | |
7JQC
 
 | | SARS-CoV-2 Nsp1, CrPV IRES and rabbit 40S ribosome complex | | 分子名称: | 40S ribosomal protein S21, 40S ribosomal protein S24, 40S ribosomal protein S26, ... | | 著者 | Yuan, S, Xiong, Y. | | 登録日 | 2020-08-10 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Nonstructural Protein 1 of SARS-CoV-2 Is a Potent Pathogenicity Factor Redirecting Host Protein Synthesis Machinery toward Viral RNA.
Mol.Cell, 80, 2020
|
|
4KXL
 
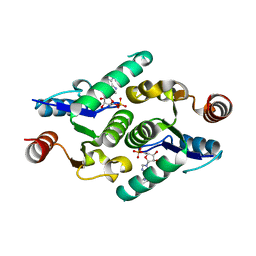 | |
3QUD
 
 | | Human p38 MAP Kinase in Complex with 2-amino-phenylamino-benzophenone | | 分子名称: | Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside, {4-[(2-aminophenyl)amino]phenyl}(phenyl)methanone | | 著者 | Gruetter, C, Rauh, D. | | 登録日 | 2011-02-23 | | 公開日 | 2012-04-11 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Resolving the selectivity problem for p38 mitogen activated protein (MAP) Kinase-inhibitors: Development of new highly potent inhibitors of p38 MAP kinase with an outstanding selectivity profile
To be Published
|
|
5NB6
 
 | | Complement factor D in complex with the inhibitor (2S,4S)-4-Amino-pyrrolidine-1,2-dicarboxylic acid 1-[(1-carbamoyl-1H-indol-3-yl)-amide] 2-[(3-trifluoromethoxy-phenyl)-amide] | | 分子名称: | (2~{S},4~{S})-~{N}1-(1-aminocarbonylindol-3-yl)-4-azanyl-~{N}2-[3-(trifluoromethyloxy)phenyl]pyrrolidine-1,2-dicarboxamide, Complement factor D | | 著者 | Mac Sweeney, A, Ostermann, N. | | 登録日 | 2017-03-01 | | 公開日 | 2017-06-28 | | 最終更新日 | 2017-07-26 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Discovery of Highly Potent and Selective Small-Molecule Reversible Factor D Inhibitors Demonstrating Alternative Complement Pathway Inhibition in Vivo.
J. Med. Chem., 60, 2017
|
|
4LIQ
 
 | | Structure of the extracellular domain of human CSF-1 receptor in complex with the Fab fragment of RG7155 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab fragment RG7155 heavy chain, Fab fragment RG7155 light chain, ... | | 著者 | Benz, J, Gorr, I.H, Hertenberger, H, Ries, C.H. | | 登録日 | 2013-07-03 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Targeting tumor-associated macrophages with anti-CSF-1R antibody reveals a strategy for cancer therapy
Cancer Cell, 25, 2014
|
|
3NWV
 
 | | Human cytochrome c G41S | | 分子名称: | Cytochrome c, HEME C | | 著者 | Fagerlund, R.D, Wilbanks, S.M. | | 登録日 | 2010-07-11 | | 公開日 | 2011-03-09 | | 最終更新日 | 2019-10-02 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The Proapoptotic G41S Mutation to Human Cytochrome c Alters the Heme Electronic Structure and Increases the Electron Self-Exchange Rate.
J.Am.Chem.Soc., 133, 2011
|
|
7JQB
 
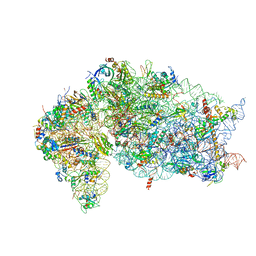 | | SARS-CoV-2 Nsp1 and rabbit 40S ribosome complex | | 分子名称: | 40S ribosomal protein S21, 40S ribosomal protein S24, 40S ribosomal protein S26, ... | | 著者 | Yuan, S, Xiong, Y. | | 登録日 | 2020-08-10 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Nonstructural Protein 1 of SARS-CoV-2 Is a Potent Pathogenicity Factor Redirecting Host Protein Synthesis Machinery toward Viral RNA.
Mol.Cell, 80, 2020
|
|
5NAT
 
 | |
4KXN
 
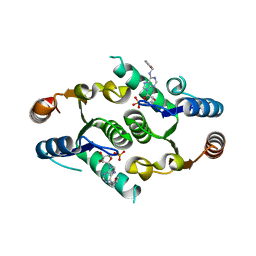 | |
4KXM
 
 | |
1FKA
 
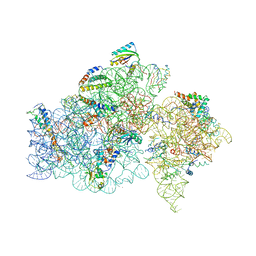 | | STRUCTURE OF FUNCTIONALLY ACTIVATED SMALL RIBOSOMAL SUBUNIT AT 3.3 A RESOLUTION | | 分子名称: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | 著者 | Schluenzen, F, Tocilj, A, Zarivach, R, Harms, J, Gluehmann, M, Janell, D, Bashan, A, Bartels, H, Agmon, I, Franceschi, F, Yonath, A. | | 登録日 | 2000-08-09 | | 公開日 | 2000-09-04 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution.
Cell(Cambridge,Mass.), 102, 2000
|
|
3QUE
 
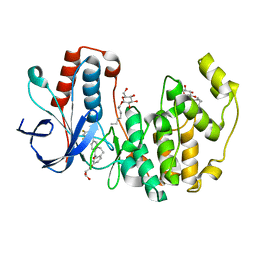 | | Human p38 MAP Kinase in Complex with Skepinone-L | | 分子名称: | 2-[(2,4-difluorophenyl)amino]-7-{[(2R)-2,3-dihydroxypropyl]oxy}-10,11-dihydro-5H-dibenzo[a,d][7]annulen-5-one, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | 著者 | Gruetter, C, Mayer-Wrangowski, S, Richters, A, Rauh, D. | | 登録日 | 2011-02-23 | | 公開日 | 2012-01-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Skepinone-L is a selective p38 mitogen-activated protein kinase inhibitor.
Nat.Chem.Biol., 8, 2012
|
|
1PMA
 
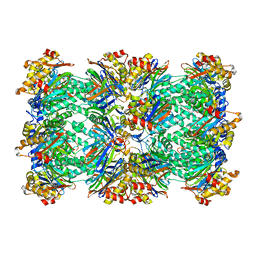 | | PROTEASOME FROM THERMOPLASMA ACIDOPHILUM | | 分子名称: | PROTEASOME | | 著者 | Loewe, J, Stock, D, Jap, B, Zwickl, P, Baumeister, W, Huber, R. | | 登録日 | 1994-12-19 | | 公開日 | 1996-06-20 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution.
Science, 268, 1995
|
|
3O98
 
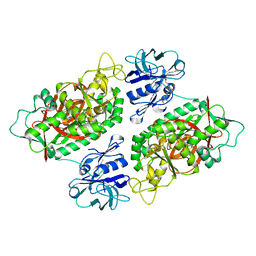 | | Glutathionylspermidine synthetase/amidase C59A complex with ADP and Gsp | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Bifunctional glutathionylspermidine synthetase/amidase, GLUTATHIONYLSPERMIDINE, ... | | 著者 | Pai, C.H, Lin, C.H, Wang, A.H.-J. | | 登録日 | 2010-08-04 | | 公開日 | 2011-03-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure and mechanism of Escherichia coli glutathionylspermidine amidase belonging to the family of cysteine; histidine-dependent amidohydrolases/peptidases
Protein Sci., 20, 2011
|
|
7L5D
 
 | | The crystal structure of SARS-CoV-2 Main Protease in complex with demethylated analog of masitinib | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Tan, K, Maltseva, N.I, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-12-21 | | 公開日 | 2020-12-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Masitinib is a broad coronavirus 3CL inhibitor that blocks replication of SARS-CoV-2.
Science, 373, 2021
|
|
2ETS
 
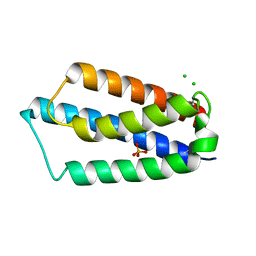 | |
3EO0
 
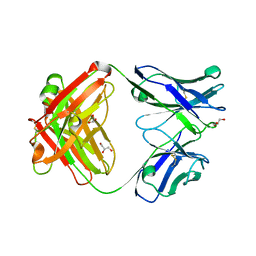 | |
2F46
 
 | |
7GAT
 
 | | SOLUTION NMR STRUCTURE OF THE L22V MUTANT DNA BINDING DOMAIN OF AREA COMPLEXED TO A 13 BP DNA CONTAINING A TGATA SITE, 34 STRUCTURES | | 分子名称: | DNA (5'-D(*CP*AP*GP*TP*GP*AP*TP*AP*GP*AP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*TP*CP*TP*AP*TP*CP*AP*CP*TP*G)-3'), NITROGEN REGULATORY PROTEIN AREA, ... | | 著者 | Clore, G.M, Starich, M, Wikstrom, M, Gronenborn, A.M. | | 登録日 | 1997-11-07 | | 公開日 | 1998-01-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of the Leu22-->Val mutant AREA DNA binding domain complexed with a TGATAG core element defines a role for hydrophobic packing in the determination of specificity.
J.Mol.Biol., 277, 1998
|
|
2F8G
 
 | | HIV-1 protease mutant I50V complexed with inhibitor TMC114 | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETIC ACID, CHLORIDE ION, ... | | 著者 | Kovalevsky, A.Y, Weber, I.T. | | 登録日 | 2005-12-02 | | 公開日 | 2006-03-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | Effectiveness of Nonpeptide Clinical Inhibitor TMC-114 on HIV-1 Protease with Highly Drug Resistant Mutations D30N, I50V, and L90M.
J.Med.Chem., 49, 2006
|
|
2F80
 
 | | HIV-1 Protease mutant D30N complexed with inhibitor TMC114 | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, POL POLYPROTEIN, ... | | 著者 | Kovalevsky, A.Y, Weber, I.T. | | 登録日 | 2005-12-01 | | 公開日 | 2006-03-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Effectiveness of Nonpeptide Clinical Inhibitor TMC-114 on HIV-1 Protease with Highly Drug Resistant Mutations D30N, I50V, and L90M.
J.Med.Chem., 49, 2006
|
|
3H0N
 
 | |
