3J8G
 
 | | Electron cryo-microscopy structure of EngA bound with the 50S ribosomal subunit | | 分子名称: | 23S rRNA, 50S ribosomal protein L1, 50S ribosomal protein L11, ... | | 著者 | Zhang, X, Yan, K, Zhang, Y, Li, N, Ma, C, Li, Z, Zhang, Y, Feng, B, Liu, J, Sun, Y, Xu, Y, Lei, J, Gao, N. | | 登録日 | 2014-10-24 | | 公開日 | 2014-11-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (5 Å) | | 主引用文献 | Structural insights into the function of a unique tandem GTPase EngA in bacterial ribosome assembly
Nucleic Acids Res., 2014
|
|
1V7Z
 
 | | creatininase-product complex | | 分子名称: | MANGANESE (II) ION, N-[(E)-AMINO(IMINO)METHYL]-N-METHYLGLYCINE, SULFATE ION, ... | | 著者 | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | 登録日 | 2003-12-26 | | 公開日 | 2004-01-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
3J3R
 
 | | Structural dynamics of the MecA-ClpC complex revealed by cryo-EM | | 分子名称: | Adapter protein MecA 1, Negative regulator of genetic competence ClpC/MecB | | 著者 | Liu, J, Mei, Z, Li, N, Qi, Y, Xu, Y, Shi, Y, Wang, F, Lei, J, Gao, N. | | 登録日 | 2013-04-18 | | 公開日 | 2013-05-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (9.4 Å) | | 主引用文献 | Structural dynamics of the MecA-ClpC complex: a type II AAA+ protein unfolding machine
J.Biol.Chem., 288, 2013
|
|
3J3U
 
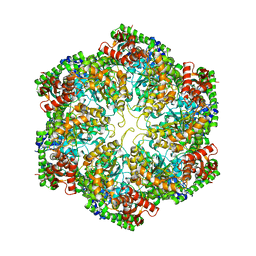 | | Structural dynamics of the MecA-ClpC complex revealed by cryo-EM | | 分子名称: | Adapter protein MecA 1, Negative regulator of genetic competence ClpC/MecB | | 著者 | Liu, J, Mei, Z, Li, N, Qi, Y, Xu, Y, Shi, Y, Wang, F, Lei, J, Gao, N. | | 登録日 | 2013-04-18 | | 公開日 | 2013-05-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (10 Å) | | 主引用文献 | Structural dynamics of the MecA-ClpC complex: a type II AAA+ protein unfolding machine.
J.Biol.Chem., 288, 2013
|
|
8QN0
 
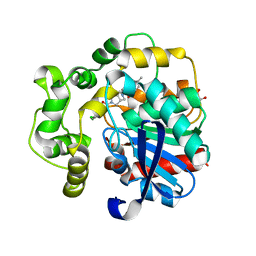 | | Soluble epoxide hydrolase in complex with RK3 | | 分子名称: | (3~{a}~{R},6~{a}~{S})-~{N}-[(2,4-dichlorophenyl)methyl]-5-(4-methylphenyl)sulfonyl-1,3,3~{a},4,6,6~{a}-hexahydropyrrolo[3,4-c]pyrrole-2-carboxamide, 1,2-ETHANEDIOL, Bifunctional epoxide hydrolase 2 | | 著者 | Kumar, A, Zhu, F, Ehrler, J.M.H, Li, F, Empel, C, Xu, Y, Atodiresei, I, Koenigs, R.M, Proschak, E, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2023-09-25 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Photosensitization enables Pauson-Khand-type reactions with nitrenes.
Science, 383, 2024
|
|
8QMZ
 
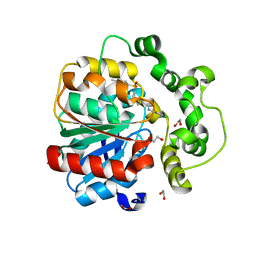 | | Soluble epoxide hydrolase in complex with RK4 | | 分子名称: | (3~{a}~{R},6~{a}~{S})-~{N}-[(2,4-dichlorophenyl)methyl]-2-(4-methylphenyl)sulfonyl-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-5-carboxamide, 1,2-ETHANEDIOL, Bifunctional epoxide hydrolase 2 | | 著者 | Kumar, A, Zhu, F, Ehrler, J.M.H, Li, F, Empel, C, Xu, Y, Atodiresei, I, Koenigs, R.M, Proschak, E, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2023-09-25 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Photosensitization enables Pauson-Khand-type reactions with nitrenes.
Science, 383, 2024
|
|
2E4I
 
 | | Human Telomeric DNA mixed-parallel/antiparallel quadruplex under Physiological Ionic Conditions Stabilized by Proper Incorporation of 8-Bromoguanosines | | 分子名称: | DNA (5'-D(*DAP*(BGM)P*DGP*DGP*DTP*DTP*DAP*(BGM)P*DGP*DGP*DTP*DTP*DAP*(BGM)P*(BGM)P*DGP*DTP*DTP*DAP*(BGM)P*DGP*DG)-3') | | 著者 | Matsugami, A, Xu, Y, Noguchi, Y, Sugiyama, H, Katahira, M. | | 登録日 | 2006-12-11 | | 公開日 | 2007-12-11 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of a human telomeric DNA sequence stabilized by 8-bromoguanosine substitutions, as determined by NMR in a K+ solution
Febs J., 274, 2007
|
|
4DUG
 
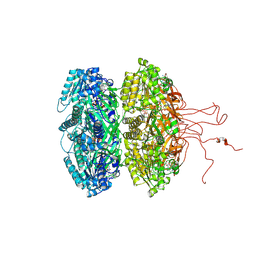 | | Crystal Structure of Circadian Clock Protein KaiC E318A Mutant | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase kaiC, MAGNESIUM ION, ... | | 著者 | Egli, M, Mori, T, Pattanayek, R, Xu, Y, Qin, X, Johnson, C.H. | | 登録日 | 2012-02-21 | | 公開日 | 2012-03-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.292 Å) | | 主引用文献 | Dephosphorylation of the Core Clock Protein KaiC in the Cyanobacterial KaiABC Circadian Oscillator Proceeds via an ATP Synthase Mechanism.
Biochemistry, 51, 2012
|
|
5COI
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | 分子名称: | 1-({[(1-ethyl-2-oxo-1,2-dihydrobenzo[cd]indol-6-yl)sulfonyl]amino}methyl)cyclopentanecarboxylic acid, Bromodomain-containing protein 4 | | 著者 | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | 登録日 | 2015-07-20 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5CPE
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N-cycloheptyl-1-ethyl-2-oxo-1,2-dihydrobenzo[cd]indole-6-sulfonamide | | 著者 | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | 登録日 | 2015-07-21 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5CP5
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | 分子名称: | 1,2-ETHANEDIOL, 1-ethyl-N-(4-fluorophenyl)-2-oxo-1,2-dihydrobenzo[cd]indole-6-sulfonamide, Bromodomain-containing protein 4, ... | | 著者 | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | 登録日 | 2015-07-21 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5CS8
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, GLYCEROL, ... | | 著者 | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | 登録日 | 2015-07-23 | | 公開日 | 2016-01-13 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5D0C
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, GLYCEROL, ... | | 著者 | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | 登録日 | 2015-08-03 | | 公開日 | 2016-01-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5CTL
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | 分子名称: | Bromodomain-containing protein 4, N-(1-ethyl-2-oxo-1,2-dihydrobenzo[cd]indol-6-yl)benzenesulfonamide | | 著者 | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | 登録日 | 2015-07-24 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5CRM
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, ... | | 著者 | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | 登録日 | 2015-07-23 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5C56
 
 | | Crystal structure of USP7/HAUSP in complex with ICP0 | | 分子名称: | Ubiquitin E3 ligase ICP0, Ubiquitin carboxyl-terminal hydrolase 7 | | 著者 | Cheng, J, Li, Z, Gong, R, Fang, J, Yang, Y, Sun, C, Yang, H, Xu, Y. | | 登録日 | 2015-06-19 | | 公開日 | 2015-07-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.685 Å) | | 主引用文献 | Molecular mechanism for the substrate recognition of USP7.
Protein Cell, 6, 2015
|
|
5D9Y
 
 | | Crystal structure of TET2-5fC complex | | 分子名称: | DNA (5'-D(*AP*CP*TP*GP*TP*(5FC)P*GP*AP*AP*GP*CP*T)-3'), DNA (5'-D(*AP*GP*CP*TP*TP*CP*GP*AP*CP*AP*GP*T)-3'), FE (III) ION, ... | | 著者 | Hu, L, Cheng, J, Rao, Q, Li, Z, Li, J, Xu, Y. | | 登録日 | 2015-08-19 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.971 Å) | | 主引用文献 | Structural insight into substrate preference for TET-mediated oxidation.
Nature, 527, 2015
|
|
5XST
 
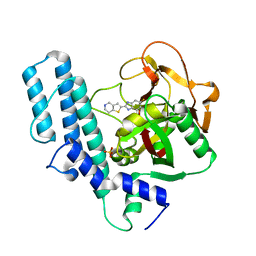 | | novel orally efficacious inhibitors complexed with PARP1 | | 分子名称: | 6-fluoranyl-2-(4,5,6,7-tetrahydrothieno[3,2-c]pyridin-2-yl)-1~{H}-benzimidazole-4-carboxamide, Poly [ADP-ribose] polymerase 1, SULFATE ION | | 著者 | Liu, Q, Xu, Y. | | 登録日 | 2017-06-15 | | 公開日 | 2018-04-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Design and synthesis of 2-(4,5,6,7-tetrahydrothienopyridin-2-yl)-benzoimidazole carboxamides as novel orally efficacious Poly(ADP-ribose)polymerase (PARP) inhibitors
Eur J Med Chem, 145, 2018
|
|
5XSR
 
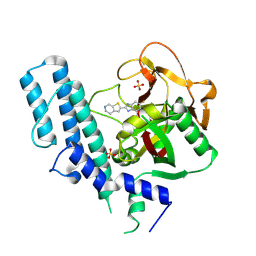 | | novel orally efficacious inhibitors complexed with PARP1 | | 分子名称: | 6-fluoranyl-2-(4,5,6,7-tetrahydrothieno[2,3-c]pyridin-2-yl)-1~{H}-benzimidazole-4-carboxamide, Poly [ADP-ribose] polymerase 1, SULFATE ION | | 著者 | Liu, Q, Xu, Y. | | 登録日 | 2017-06-15 | | 公開日 | 2018-04-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Design and synthesis of 2-(4,5,6,7-tetrahydrothienopyridin-2-yl)-benzoimidazole carboxamides as novel orally efficacious Poly(ADP-ribose)polymerase (PARP) inhibitors
Eur J Med Chem, 145, 2018
|
|
5Y3R
 
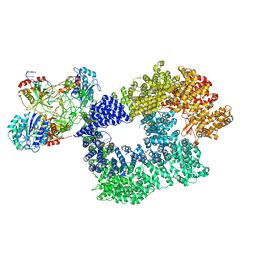 | | Cryo-EM structure of Human DNA-PK Holoenzyme | | 分子名称: | DNA (34-MER), DNA (36-MER), DNA-dependent protein kinase catalytic subunit, ... | | 著者 | Yin, X, Liu, M, Tian, Y, Wang, J, Xu, Y. | | 登録日 | 2017-07-29 | | 公開日 | 2017-09-06 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (6.6 Å) | | 主引用文献 | Cryo-EM structure of human DNA-PK holoenzyme
Cell Res., 27, 2017
|
|
5Y9Q
 
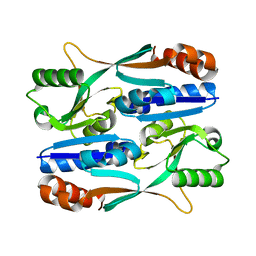 | |
5YA5
 
 | | CRYSTAL STRUCTURE OF c-MET IN COMPLEX WITH NOVEL INHIBITOR | | 分子名称: | 2-[3-(4-methoxybenzyl)[1,2,4]triazolo[3,4-b][1,3,4]thiadiazol-6-yl]-1H-indole, Hepatocyte growth factor receptor | | 著者 | Liu, Q, Xu, Y. | | 登録日 | 2017-08-30 | | 公開日 | 2018-07-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Discovery, optimization and biological evaluation for novel c-Met kinase inhibitors
Eur J Med Chem, 143, 2018
|
|
7MYO
 
 | | Cryo-EM structure of p110alpha in complex with p85alpha inhibited by BYL-719 | | 分子名称: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | 著者 | Liu, X, Yang, S, Hart, J.R, Xu, Y, Zou, X, Zhang, H, Zhou, Q, Xia, T, Zhang, Y, Yang, D, Wang, M.-W, Vogt, P.K. | | 登録日 | 2021-05-21 | | 公開日 | 2021-11-10 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.92 Å) | | 主引用文献 | Cryo-EM structures of PI3K alpha reveal conformational changes during inhibition and activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MYN
 
 | | Cryo-EM Structure of p110alpha in complex with p85alpha | | 分子名称: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | 著者 | Liu, X, Yang, S, Hart, J.R, Xu, Y, Zou, X, Zhang, H, Zhou, Q, Xia, T, Zhang, Y, Yang, D, Wang, M.-W, Vogt, P.K. | | 登録日 | 2021-05-21 | | 公開日 | 2021-11-10 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.79 Å) | | 主引用文献 | Cryo-EM structures of PI3K alpha reveal conformational changes during inhibition and activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8WJW
 
 | |
