7TO4
 
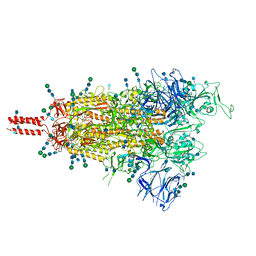 | | Structural and functional impact by SARS-CoV-2 Omicron spike mutations | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | 登録日 | 2022-01-22 | | 公開日 | 2022-02-16 | | 最終更新日 | 2022-05-11 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural and functional impact by SARS-CoV-2 Omicron spike mutations.
Cell Rep, 39, 2022
|
|
2KH5
 
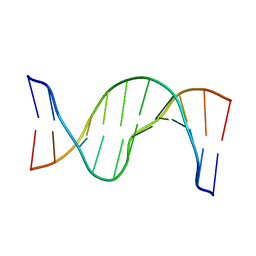 | | Solution Structure of cis-5R,6S-thymine glycol opposite complementary adenine in duplex DNA | | 分子名称: | 5'-D(*AP*CP*AP*AP*AP*CP*AP*CP*GP*CP*AP*C)-3', 5'-D(*GP*TP*GP*CP*GP*(CTG)P*GP*TP*TP*TP*GP*T)-3' | | 著者 | Brown, K.L. | | 登録日 | 2009-03-24 | | 公開日 | 2010-03-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Binding of the human nucleotide excision repair proteins XPA and XPC/HR23B to the 5R-thymine glycol lesion and structure of the cis-(5R,6S) thymine glycol epimer in the 5'-GTgG-3' sequence: destabilization of two base pairs at the lesion site
Nucleic Acids Res., 38, 2010
|
|
4BJM
 
 | |
4BJN
 
 | |
6AKG
 
 | |
6AKF
 
 | |
6AKE
 
 | |
8K5Y
 
 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | 分子名称: | (3-azanyl-4-fluoranyl-5,7,8,9-tetrahydropyrido[4,3-c]azepin-6-yl)-[6-(2-oxidanylpropan-2-yl)-1H-indol-2-yl]methanone, CALCIUM ION, DIHYDROGENPHOSPHATE ION, ... | | 著者 | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | 登録日 | 2023-07-24 | | 公開日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
8K5V
 
 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | 分子名称: | 6,7-dihydro-4H-[1,3]oxazolo[4,5-c]pyridin-5-yl-(7-ethyl-2H-indazol-3-yl)methanone, CALCIUM ION, DIHYDROGENPHOSPHATE ION, ... | | 著者 | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | 登録日 | 2023-07-24 | | 公開日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
8K5X
 
 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | 分子名称: | (6-cyclopropyl-1~{H}-indol-2-yl)-(5,7,8,9-tetrahydropyrido[4,3-c]azepin-6-yl)methanone, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | 登録日 | 2023-07-24 | | 公開日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
7F5G
 
 | | The crystal structure of RBD-Nanobody complex, DL4 (SA4) | | 分子名称: | ACETATE ION, GLYCEROL, Nanobody DL4, ... | | 著者 | Li, T, Lai, Y, Zhou, Y, Tan, J, Li, D. | | 登録日 | 2021-06-22 | | 公開日 | 2022-05-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Isolation, characterization, and structure-based engineering of a neutralizing nanobody against SARS-CoV-2.
Int.J.Biol.Macromol., 209, 2022
|
|
5I7U
 
 | | Human DPP4 in complex with a novel tricyclic hetero-cycle inhibitor | | 分子名称: | 2-({2-[(3R)-3-aminopiperidin-1-yl]-5-methyl-6,9-dioxo-5,6,7,9-tetrahydro-1H-imidazo[1,2-a]purin-1-yl}methyl)-4-fluorobenzonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Scapin, G. | | 登録日 | 2016-02-18 | | 公開日 | 2016-06-08 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Discovery of Novel Tricyclic Heterocycles as Potent and Selective DPP-4 Inhibitors for the Treatment of Type 2 Diabetes.
Acs Med.Chem.Lett., 7, 2016
|
|
7F5H
 
 | | The crystal structure of RBD-Nanobody complex, DL28 (SC4) | | 分子名称: | GLYCEROL, Nanobody DL28, PHOSPHATE ION, ... | | 著者 | Luo, Z.P, Li, T, Lai, Y, Zhou, Y, Tan, J, Li, D. | | 登録日 | 2021-06-22 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Characterization of a Neutralizing Nanobody With Broad Activity Against SARS-CoV-2 Variants.
Front Microbiol, 13, 2022
|
|
7KMY
 
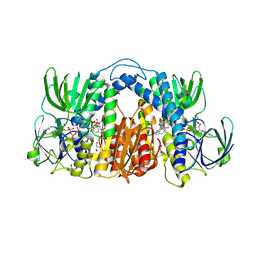 | | Structure of Mtb Lpd bound to 010705 | | 分子名称: | Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | 著者 | Lima, C.D. | | 登録日 | 2020-11-03 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Whole Cell Active Inhibitors of Mycobacterial Lipoamide Dehydrogenase Afford Selectivity over the Human Enzyme through Tight Binding Interactions.
Acs Infect Dis., 7, 2021
|
|
2BV8
 
 | | The crystal structure of Phycocyanin from Gracilaria chilensis. | | 分子名称: | C-PHYCOCYANIN ALPHA SUBUNIT, C-PHYCOCYANIN BETA SUBUNIT, PHYCOCYANOBILIN, ... | | 著者 | Contreras-Martel, C, Martinez-Oyanedel, J, Poo-Caama, G, Bruna, C, Bunster, M. | | 登録日 | 2005-06-23 | | 公開日 | 2006-08-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | The Structure at 2 A Resolution of Phycocyanin from Gracilaria Chilensis and the Energy Transfer Network in a Pc-Pc Complex.
Biophys.Chem., 125, 2007
|
|
3X1O
 
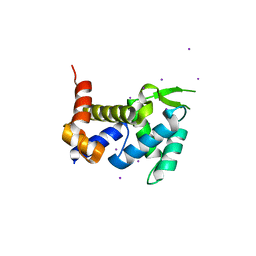 | | Crystal structure of the ROQ domain of human Roquin | | 分子名称: | IODIDE ION, Roquin-1 | | 著者 | Ose, T, Verma, A, Cockburn, J.B, Berrow, N.S, Alderton, D, Stuart, D, Owens, R.J, Jones, E.Y. | | 登録日 | 2014-11-26 | | 公開日 | 2015-03-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.201 Å) | | 主引用文献 | Roquin binds microRNA-146a and Argonaute2 to regulate microRNA homeostasis
Nat Commun, 6, 2015
|
|
6CU7
 
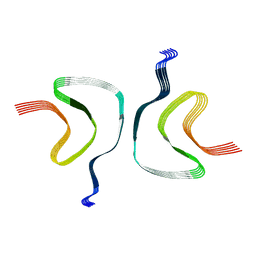 | | Alpha Synuclein fibril formed by full length protein - Rod Polymorph | | 分子名称: | Alpha-synuclein | | 著者 | Li, B, Hatami, A, Ge, P, Murray, K.A, Sheth, P, Zhang, M, Nair, G, Sawaya, M.R, Zhu, C, Broad, M, Shin, W.S, Ye, S, John, V, Eisenberg, D.S, Zhou, Z.H, Jiang, L. | | 登録日 | 2018-03-23 | | 公開日 | 2018-09-12 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Cryo-EM of full-length alpha-synuclein reveals fibril polymorphs with a common structural kernel.
Nat Commun, 9, 2018
|
|
6CU8
 
 | | Alpha Synuclein fibril formed by full length protein - Twister Polymorph | | 分子名称: | Alpha-synuclein | | 著者 | Li, B, Hatami, A, Ge, P, Murray, K.A, Sheth, P, Zhang, M, Nair, G, Sawaya, M.R, Zhu, C, Broad, M, Shin, W.S, Ye, S, John, V, Eisenberg, D.S, Zhou, Z.H, Jiang, L. | | 登録日 | 2018-03-23 | | 公開日 | 2018-09-12 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM of full-length alpha-synuclein reveals fibril polymorphs with a common structural kernel.
Nat Commun, 9, 2018
|
|
6I8G
 
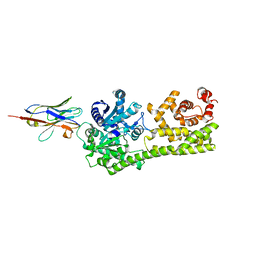 | |
4MPA
 
 | | Crystal structure of NHERF1-CXCR2 signaling complex in P21 space group | | 分子名称: | ACETIC ACID, CHLORIDE ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1, ... | | 著者 | Jiang, Y, Lu, G, Wu, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | 登録日 | 2013-09-12 | | 公開日 | 2014-01-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.097 Å) | | 主引用文献 | New Conformational State of NHERF1-CXCR2 Signaling Complex Captured by Crystal Lattice Trapping.
Plos One, 8, 2013
|
|
6I8H
 
 | |
4LMM
 
 | | Crystal structure of NHERF1 PDZ1 domain complexed with the CXCR2 C-terminal tail in P21 space group | | 分子名称: | ACETIC ACID, CHLORIDE ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | 著者 | Jiang, Y, Lu, G, Wu, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | 登録日 | 2013-07-10 | | 公開日 | 2014-01-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | New Conformational State of NHERF1-CXCR2 Signaling Complex Captured by Crystal Lattice Trapping.
Plos One, 8, 2013
|
|
4R91
 
 | | BACE-1 in complex with (R)-4-(2-cyclohexylethyl)-4-(((1S,3R)-3-(cyclopentylamino)cyclohexyl)methyl)-1-methyl-5-oxoimidazolidin-2-iminium | | 分子名称: | (2E,5R)-5-(2-cyclohexylethyl)-5-{[(1S,3R)-3-(cyclopentylamino)cyclohexyl]methyl}-2-imino-3-methylimidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | 著者 | Orth, P, Caldwell, J.P, Strickland, C. | | 登録日 | 2014-09-03 | | 公開日 | 2014-11-05 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4R92
 
 | | BACE-1 in complex with (R)-4-(2-cyclohexylethyl)-4-(((1S,3R)-3-(isonicotinamido)cyclohexyl)methyl)-1-methyl-5-oxoimidazolidin-2-iminium | | 分子名称: | Beta-secretase 1, L(+)-TARTARIC ACID, N-[(1R,3S)-3-{[(2E,4R)-4-(2-cyclohexylethyl)-2-imino-1-methyl-5-oxoimidazolidin-4-yl]methyl}cyclohexyl]pyridine-4-carboxamide | | 著者 | Orth, P, Strickland, C, Caldwell, J.P. | | 登録日 | 2014-09-03 | | 公開日 | 2014-11-05 | | 最終更新日 | 2014-12-17 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4R95
 
 | | BACE-1 in complex with 2-(((1R,3S)-3-(((R)-4-(2-cyclohexylethyl)-2-iminio-1-methyl-5-oxoimidazolidin-4-yl)methyl)cyclohexyl)amino)quinolin-1-ium | | 分子名称: | (2E,5R)-5-(2-cyclohexylethyl)-2-imino-3-methyl-5-{[(1S,3R)-3-(quinolin-2-ylamino)cyclohexyl]methyl}imidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | 著者 | Orth, P, Strickland, C, Caldwell, J.P. | | 登録日 | 2014-09-03 | | 公開日 | 2014-11-05 | | 最終更新日 | 2014-12-17 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
