2E80
 
 | |
2FT6
 
 | |
2FT8
 
 | |
2FT7
 
 | |
2FTA
 
 | |
6R26
 
 | | The photosensory core module (PAS-GAF-PHY) of the bacterial phytochrome Agp1 (AtBphP1) locked in a Pr-like state | | 分子名称: | 3-[2-[(~{Z})-[12-ethyl-6-(3-hydroxy-3-oxopropyl)-13-methyl-11-oxidanylidene-4,10-diazatricyclo[8.3.0.0^{3,7}]trideca-1,3,6,12-tetraen-5-ylidene]methyl]-5-[(~{Z})-(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Bacteriophytochrome protein, CALCIUM ION | | 著者 | Scheerer, P, Michael, N, Lamparter, T, Krauss, N. | | 登録日 | 2019-03-15 | | 公開日 | 2020-04-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.11 Å) | | 主引用文献 | Crystal structures of the photosensory core module of bacteriophytochrome Agp1 reveal pronounced structural flexibility of this protein in the red-absorbing Pr state
To Be Published
|
|
6R27
 
 | | Crystallographic superstructure of the photosensory core module (PAS-GAF-PHY) of the bacterial phytochrome Agp1 (AtBphP1) locked in a Pr-like state | | 分子名称: | 3-[2-[(~{Z})-[12-ethyl-6-(3-hydroxy-3-oxopropyl)-13-methyl-11-oxidanylidene-4,10-diazatricyclo[8.3.0.0^{3,7}]trideca-1,3,6,12-tetraen-5-ylidene]methyl]-5-[(~{Z})-(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Bacteriophytochrome protein | | 著者 | Scheerer, P, Michael, N, Lamparter, T, Krauss, N. | | 登録日 | 2019-03-15 | | 公開日 | 2020-04-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Crystal structures of the photosensory core module of bacteriophytochrome Agp1 reveal pronounced structural flexibility of this protein in the red-absorbing Pr state
To Be Published
|
|
5XRX
 
 | | EFK17DA structure in Microgel MAA60 | | 分子名称: | Cathelicidin antimicrobial peptide | | 著者 | Datta, A, Bhunia, A. | | 登録日 | 2017-06-10 | | 公開日 | 2018-04-18 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Conformational Aspects of High Content Packing of Antimicrobial Peptides in Polymer Microgels
ACS Appl Mater Interfaces, 9, 2017
|
|
8C3U
 
 | | Crystal Structure of human IL-1beta in complex with a low molecular weight antagonist | | 分子名称: | (S)-4'-hydroxy-3'-(6-methyl-2-oxo-3-(1H-pyrazol-4-yl)indolin-3-yl)-[1,1'-biphenyl]-2,4-dicarboxylic acid, Interleukin-1 beta | | 著者 | Rondeau, J.-M, Lehmann, S, Koch, E. | | 登録日 | 2022-12-28 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.945 Å) | | 主引用文献 | Discovery of a selective and biologically active low-molecular weight antagonist of human interleukin-1 beta.
Nat Commun, 14, 2023
|
|
1GC2
 
 | | CRYSTAL STRUCTURE OF THE PYRIDOXAL-5'-PHOSPHATE DEPENDENT L-METHIONINE GAMMA-LYASE FROM PSEUDOMONAS PUTIDA | | 分子名称: | METHIONINE GAMMA-LYASE | | 著者 | Motoshima, H, Inagaki, K, Kumasaka, T, Furuichi, M, Inoue, H, Tamura, T, Esaki, N, Soda, K, Tanaka, N, Yamamoto, M, Tanaka, H. | | 登録日 | 2000-07-06 | | 公開日 | 2002-05-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the pyridoxal 5'-phosphate dependent L-methionine gamma-lyase from Pseudomonas putida.
J.Biochem., 128, 2000
|
|
2L7U
 
 | | Structure of CEL-PEP-RAGE V domain complex | | 分子名称: | Advanced glycosylation end product-specific receptor, Serum albumin peptide | | 著者 | Xue, J, Rai, V, Schmidt, A, Frolov, S, Reverdatto, S, Singer, D, Chabierski, S, Xie, J, Burz, D, Shekhtman, A, Hoffman, R. | | 登録日 | 2010-12-23 | | 公開日 | 2011-05-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Advanced glycation end product recognition by the receptor for AGEs.
Structure, 19, 2011
|
|
2LMB
 
 | | Solution Structure of C-terminal RAGE (ctRAGE) | | 分子名称: | Advanced glycosylation end product-specific receptor | | 著者 | Rai, V, Maldonado, A.Y, Burz, D.S, Reverdatto, S, Schmidt, A, Shekhtman, A. | | 登録日 | 2011-11-29 | | 公開日 | 2011-12-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Signal transduction in receptor for advanced glycation end products (RAGE): solution structure of C-terminal rage (ctRAGE) and its binding to mDia1.
J.Biol.Chem., 287, 2012
|
|
1GC0
 
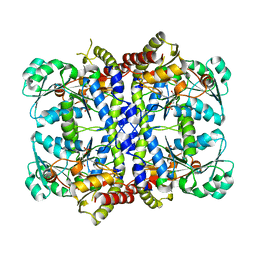 | | CRYSTAL STRUCTURE OF THE PYRIDOXAL-5'-PHOSPHATE DEPENDENT L-METHIONINE GAMMA-LYASE FROM PSEUDOMONAS PUTIDA | | 分子名称: | METHIONINE GAMMA-LYASE | | 著者 | Motoshima, H, Inagaki, K, Kumasaka, T, Furuichi, M, Inoue, H, Tamura, T, Esaki, N, Soda, K, Tanaka, N, Yamamoto, M, Tanaka, H. | | 登録日 | 2000-07-06 | | 公開日 | 2002-05-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of the pyridoxal 5'-phosphate dependent L-methionine gamma-lyase from Pseudomonas putida.
J.Biochem., 128, 2000
|
|
8RYS
 
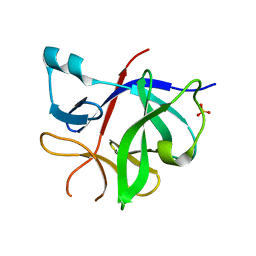 | | Human IL-1beta, unliganded | | 分子名称: | Interleukin-1 beta, SULFATE ION | | 著者 | Rondeau, J.-M, Lehmann, S. | | 登録日 | 2024-02-09 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Ligandability Assessment of IL-1 beta by Integrated Hit Identification Approaches.
J.Med.Chem., 67, 2024
|
|
8RYK
 
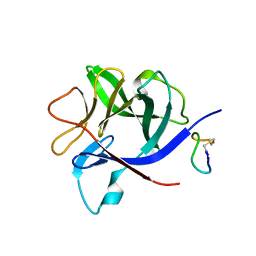 | |
8RZB
 
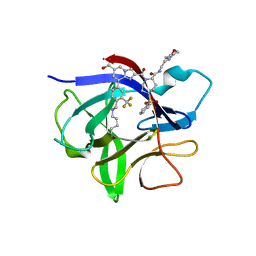 | | IL-1beta in complex with covalent DEL hit | | 分子名称: | 8-[4-methyl-3-(trifluoromethyl)phenyl]-2-[[(7S)-7-(2-morpholin-4-ylethylcarbamoyl)-4-(phenylsulfonyl)-1,4-diazepan-1-yl]carbonyl]imidazo[1,2-a]pyridine-6-carboxylic acid, Interleukin-1 beta | | 著者 | Rondeau, J.-M, Lehmann, S. | | 登録日 | 2024-02-12 | | 公開日 | 2024-05-22 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.836 Å) | | 主引用文献 | Ligandability Assessment of IL-1 beta by Integrated Hit Identification Approaches.
J.Med.Chem., 67, 2024
|
|
5TRX
 
 | | Room temperature structure of an extradiol ring-cleaving dioxygenase from B.fuscum determined using serial femtosecond crystallography | | 分子名称: | CALCIUM ION, CHLORIDE ION, FE (II) ION, ... | | 著者 | Kovaleva, E.G, Oberthuer, D, Tolstikova, A, Mariani, V. | | 登録日 | 2016-10-27 | | 公開日 | 2017-03-22 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Double-flow focused liquid injector for efficient serial femtosecond crystallography
Sci. Rep., 7, 2017
|
|
6G9P
 
 | |
6H1E
 
 | | Crystal structure of C21orf127-TRMT112 in complex with SAH and H4 peptide | | 分子名称: | HemK methyltransferase family member 2, Histone H4 peptide, Multifunctional methyltransferase subunit TRM112-like protein, ... | | 著者 | Wang, S, Hermann, B, Metzger, E, Peng, L, Einsle, O, Schuele, R. | | 登録日 | 2018-07-11 | | 公開日 | 2019-05-22 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | KMT9 monomethylates histone H4 lysine 12 and controls proliferation of prostate cancer cells.
Nat.Struct.Mol.Biol., 26, 2019
|
|
5U5Q
 
 | | 12 Subunit RNA Polymerase II at Room Temperature collected using SFX | | 分子名称: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | 著者 | Bushnell, D.A, Oberthur, D, Mariani, V, Yefanov, O, Tolstikova, A, Barty, A. | | 登録日 | 2016-12-07 | | 公開日 | 2017-03-29 | | 最終更新日 | 2019-11-27 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Double-flow focused liquid injector for efficient serial femtosecond crystallography.
Sci Rep, 7, 2017
|
|
6H1D
 
 | | Crystal structure of C21orf127-TRMT112 in complex with SAH | | 分子名称: | HemK methyltransferase family member 2, Multifunctional methyltransferase subunit TRM112-like protein, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Wang, S, Hermann, B, Metzger, E, Peng, L, Einsle, O, Schuele, R. | | 登録日 | 2018-07-11 | | 公開日 | 2019-05-22 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | KMT9 monomethylates histone H4 lysine 12 and controls proliferation of prostate cancer cells.
Nat.Struct.Mol.Biol., 26, 2019
|
|
8OUA
 
 | | Cereblon isoform 4 in complex with novel Benzamide-Type Cereblon Binder 11f | | 分子名称: | 4-azanyl-~{N}-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-2,5-bis(fluoranyl)-3-methoxy-benzamide, Cereblon isoform 4, PHOSPHATE ION, ... | | 著者 | Heim, C, Bischof, L, Hartmann, M.D. | | 登録日 | 2023-04-22 | | 公開日 | 2023-11-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Leveraging Ligand Affinity and Properties: Discovery of Novel Benzamide-Type Cereblon Binders for the Design of PROTACs.
J.Med.Chem., 66, 2023
|
|
8OU4
 
 | | Cereblon isoform 4 in complex with novel Benzamide-Type Cereblon Binder 11a | | 分子名称: | 4-azanyl-~{N}-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-2-chloranyl-benzamide, Cereblon isoform 4, ZINC ION | | 著者 | Heim, C, Bischof, L, Hartmann, M.D. | | 登録日 | 2023-04-21 | | 公開日 | 2023-11-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Leveraging Ligand Affinity and Properties: Discovery of Novel Benzamide-Type Cereblon Binders for the Design of PROTACs.
J.Med.Chem., 66, 2023
|
|
8OU6
 
 | | Cereblon isoform 4 in complex with novel Benzamide-Type Cereblon Binder 11c | | 分子名称: | 4-azanyl-~{N}-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-2-methyl-benzamide, Cereblon isoform 4, PHOSPHATE ION, ... | | 著者 | Heim, C, Bischof, L, Hartmann, M.D. | | 登録日 | 2023-04-21 | | 公開日 | 2023-11-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Leveraging Ligand Affinity and Properties: Discovery of Novel Benzamide-Type Cereblon Binders for the Design of PROTACs.
J.Med.Chem., 66, 2023
|
|
8OU3
 
 | | Cereblon isoform 4 in complex with novel Benzamide-Type Cereblon Binder 8d | | 分子名称: | 4-azanyl-~{N}-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-2-fluoranyl-benzamide, Cereblon isoform 4, PHOSPHATE ION, ... | | 著者 | Heim, C, Bischof, L, Maiwald, S, Hartmann, M.D. | | 登録日 | 2023-04-21 | | 公開日 | 2023-11-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Leveraging Ligand Affinity and Properties: Discovery of Novel Benzamide-Type Cereblon Binders for the Design of PROTACs.
J.Med.Chem., 66, 2023
|
|
