6CDY
 
 | | Crystal structure of TEAD complexed with its inhibitor | | 分子名称: | 2-[(4H-1,2,4-triazol-3-yl)sulfanyl]-N-{4-[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]decan-1-yl]phenyl}acetamide, Transcriptional enhancer factor TEF-4 | | 著者 | LIU, S, HAN, X, LUO, X. | | 登録日 | 2018-02-09 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Lats1/2 Sustain Intestinal Stem Cells and Wnt Activation through TEAD-Dependent and Independent Transcription.
Cell Stem Cell, 26, 2020
|
|
6CUC
 
 | |
2LP6
 
 | | Refined Solution NMR Structure of the 50S ribosomal protein L35Ae from Pyrococcus furiosus, Northeast Structural Genomics Consortium Target (NESG) PfR48 | | 分子名称: | 50S ribosomal protein L35Ae | | 著者 | Snyder, D.A, Aramini, J.M, Yu, B, Huang, Y.J, Xiao, R, Cort, J.R, Shastry, R, Ma, L, Liu, J, Rost, B, Acton, T.B, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-02-02 | | 公開日 | 2012-02-15 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure of the ribosomal protein RP-L35Ae from Pyrococcus furiosus.
Proteins, 80, 2012
|
|
9FQI
 
 | | E3 ligase Cbl-b in complex with a lactam scaffold inhibitor (compound 7) | | 分子名称: | 8-[3-[(4~{R})-4-methyl-2-oxidanylidene-piperidin-4-yl]phenyl]-3-[[(3~{S})-3-methylpiperidin-1-yl]methyl]-5-(trifluoromethyl)-1$l^{4},7,8-triazabicyclo[4.3.0]nona-1(6),2,4-trien-9-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | 著者 | Schimpl, M. | | 登録日 | 2024-06-17 | | 公開日 | 2024-07-31 | | 最終更新日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (1.954 Å) | | 主引用文献 | Accelerated Discovery of Carbamate Cbl-b Inhibitors Using Generative AI Models and Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
9FQJ
 
 | | E3 ligase Cbl-b in complex with a carbamate scaffold inhibitor (compound 12) | | 分子名称: | 2-cyclopropyl-6-methyl-~{N}-[3-[(6~{S})-6-methyl-2-oxidanylidene-1,3-oxazinan-6-yl]phenyl]pyrimidine-4-carboxamide, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | 著者 | Schimpl, M. | | 登録日 | 2024-06-17 | | 公開日 | 2024-07-31 | | 最終更新日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (1.563 Å) | | 主引用文献 | Accelerated Discovery of Carbamate Cbl-b Inhibitors Using Generative AI Models and Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
9FQH
 
 | | E3 ligase Cbl-b in complex with a triazolone core inhibitor (compound 1) | | 分子名称: | 8-[3-[3-methyl-1-(4-methyl-1,2,4-triazol-3-yl)cyclobutyl]phenyl]-3-[[(3~{S})-3-methylpiperidin-1-yl]methyl]-5-(trifluoromethyl)-1$l^{4},7,8-triazabicyclo[4.3.0]nona-1(6),2,4-trien-9-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | 著者 | Schimpl, M. | | 登録日 | 2024-06-17 | | 公開日 | 2024-07-31 | | 最終更新日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (1.786 Å) | | 主引用文献 | Accelerated Discovery of Carbamate Cbl-b Inhibitors Using Generative AI Models and Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
7BR3
 
 | | Crystal structure of the protein 1 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R)-2,3-dihydroxypropyl dodecanoate, 4-[[(1R)-2-[5-(2-fluoranyl-3-methoxy-phenyl)-3-[[2-fluoranyl-6-(trifluoromethyl)phenyl]methyl]-4-methyl-2,6-bis(oxidanylidene)pyrimidin-1-yl]-1-phenyl-ethyl]amino]butanoic acid, ... | | 著者 | Cheng, L, Shao, Z. | | 登録日 | 2020-03-26 | | 公開日 | 2020-10-07 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Structure of the human gonadotropin-releasing hormone receptor GnRH1R reveals an unusual ligand binding mode.
Nat Commun, 11, 2020
|
|
4XNW
 
 | | The human P2Y1 receptor in complex with MRS2500 | | 分子名称: | P2Y purinoceptor 1,Rubredoxin,P2Y purinoceptor 1, ZINC ION, [(1R,2S,4S,5S)-4-[2-iodo-6-(methylamino)-9H-purin-9-yl]-2-(phosphonooxy)bicyclo[3.1.0]hex-1-yl]methyl dihydrogen phosphate | | 著者 | Zhang, D, Gao, Z, Jacobson, K, Han, G.W, Stevens, R, Zhao, Q, Wu, B, GPCR Network (GPCR) | | 登録日 | 2015-01-16 | | 公開日 | 2015-04-01 | | 最終更新日 | 2020-02-05 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Two disparate ligand-binding sites in the human P2Y1 receptor
Nature, 520, 2015
|
|
4XNV
 
 | | The human P2Y1 receptor in complex with BPTU | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2-(2-tert-butylphenoxy)pyridin-3-yl]-3-[4-(trifluoromethoxy)phenyl]urea, CHOLESTEROL, ... | | 著者 | Zhang, D, Gao, Z, Jacobson, K, Han, G.W, Stevens, R, Zhao, Q, Wu, B, GPCR Network (GPCR) | | 登録日 | 2015-01-16 | | 公開日 | 2015-04-01 | | 最終更新日 | 2020-02-05 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Two disparate ligand-binding sites in the human P2Y1 receptor
Nature, 520, 2015
|
|
4YOC
 
 | | Crystal Structure of human DNMT1 and USP7/HAUSP complex | | 分子名称: | DNA (cytosine-5)-methyltransferase 1, Ubiquitin carboxyl-terminal hydrolase 7, ZINC ION | | 著者 | Cheng, J, Yang, H, Fang, J, Gong, R, Wang, P, Li, Z, Xu, Y. | | 登録日 | 2015-03-11 | | 公開日 | 2015-05-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.916 Å) | | 主引用文献 | Molecular mechanism for USP7-mediated DNMT1 stabilization by acetylation.
Nat Commun, 6, 2015
|
|
4ZT9
 
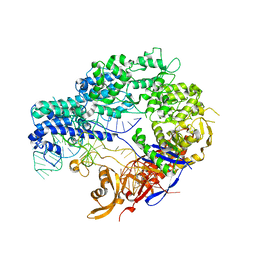 | |
4ZT0
 
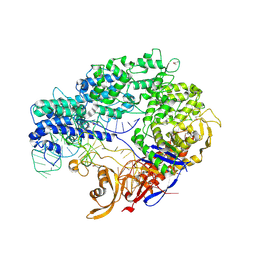 | |
2JRT
 
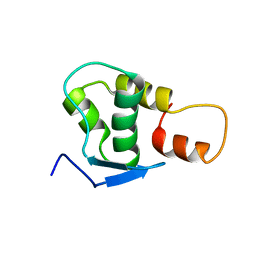 | | NMR solution structure of the protein coded by gene RHOS4_12090 of Rhodobacter sphaeroides. Northeast Structural Genomics target RhR5 | | 分子名称: | Uncharacterized protein | | 著者 | Wang, L, Chen, C, Nwosu, C, Cunningham, K, Owens, L, Ma, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2007-06-28 | | 公開日 | 2007-08-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of the protein coded by gene RHOS4_12090 of Rhodobacter sphaeroides.
To be Published
|
|
7CK8
 
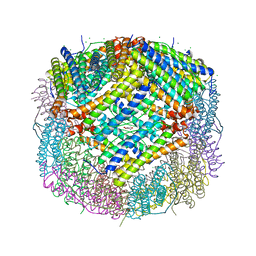 | | Crystal structure of human ferritin heavy chain mutant C90S/C102S/C130S | | 分子名称: | CHLORIDE ION, FE (III) ION, Ferritin heavy chain, ... | | 著者 | Chen, X, Jiang, B, Yan, X, Fan, K. | | 登録日 | 2020-07-16 | | 公開日 | 2021-05-26 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A natural drug entry channel in the ferritin nanocage.
Nano Today, 35, 2020
|
|
7CMA
 
 | | Structure of A151R from African swine fever virus Georgia | | 分子名称: | A151R, ZINC ION | | 著者 | Niu, D, Liu, K, Huang, J, Chen, C, Liu, W, Guo, R. | | 登録日 | 2020-07-26 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structure basis of non-structural protein pA151R from African Swine Fever Virus.
Biochem.Biophys.Res.Commun., 532, 2020
|
|
7DC6
 
 | | Giant panda MHC class I complexes | | 分子名称: | Beta-2-microglobulin, CCV-NGY9 peptide from Spike protein, MHC class I antigen | | 著者 | Yuan, H, Xia, C. | | 登録日 | 2020-10-23 | | 公開日 | 2021-06-16 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Crystal structure of the giant panda MHC class I complex: First insights into the viral peptide presentation profile in the bear family.
Protein Sci., 29, 2020
|
|
5LMZ
 
 | | Fluorinase from Streptomyces sp. MA37 | | 分子名称: | 1-DEAZA-ADENOSINE, CHLORIDE ION, Fluorinase, ... | | 著者 | Mann, G, O'Hagan, D, Naismith, J.H, Bandaranayaka, N. | | 登録日 | 2016-08-02 | | 公開日 | 2016-08-24 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Identification of fluorinases from Streptomyces sp MA37, Norcardia brasiliensis, and Actinoplanes sp N902-109 by genome mining.
Chembiochem, 15, 2014
|
|
4FLP
 
 | | Crystal Structure of the first bromodomain of human BRDT in complex with the inhibitor JQ1 | | 分子名称: | (6S)-6-(2-tert-butoxy-2-oxoethyl)-4-(4-chlorophenyl)-2,3,9-trimethyl-6,7-dihydrothieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-10-ium, Bromodomain testis-specific protein, POTASSIUM ION | | 著者 | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, Canning, P, Muniz, J, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Bradner, J, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2012-06-15 | | 公開日 | 2012-07-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Small-Molecule Inhibition of BRDT for Male Contraception.
Cell(Cambridge,Mass.), 150, 2012
|
|
8EWG
 
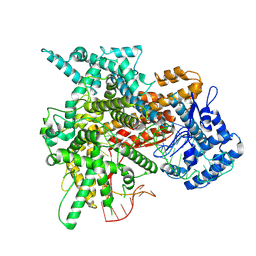 | | Cryo-EM structure of a riboendonclease | | 分子名称: | CRISPR-associated endonuclease Cas9, RNA (56-MER) | | 著者 | Li, Z, Wang, F. | | 登録日 | 2022-10-23 | | 公開日 | 2023-08-30 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural Basis for the Ribonuclease Activity of a Thermostable CRISPR-Cas13a from Thermoclostridium caenicola.
J.Mol.Biol., 435, 2023
|
|
4MBS
 
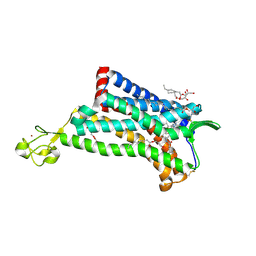 | | Crystal Structure of the CCR5 Chemokine Receptor | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4,4-difluoro-N-[(1S)-3-{(3-exo)-3-[3-methyl-5-(propan-2-yl)-4H-1,2,4-triazol-4-yl]-8-azabicyclo[3.2.1]oct-8-yl}-1-phenylpropyl]cyclohexanecarboxamide, Chimera protein of C-C chemokine receptor type 5 and Rubredoxin, ... | | 著者 | Tan, Q, Zhu, Y, Han, G.W, Li, J, Fenalti, G, Liu, H, Cherezov, V, Stevens, R.C, GPCR Network (GPCR), Zhao, Q, Wu, B. | | 登録日 | 2013-08-19 | | 公開日 | 2013-09-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex.
Science, 341, 2013
|
|
6LW5
 
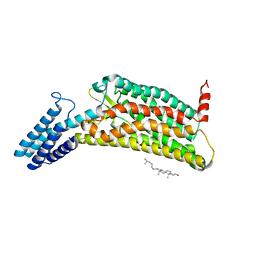 | | Crystal structure of the human formyl peptide receptor 2 in complex with WKYMVm | | 分子名称: | CHOLESTEROL, Soluble cytochrome b562,N-formyl peptide receptor 2, TRP-LYS-TYR-MET-VAL-QXV | | 著者 | Chen, T, Zong, X, Zhang, H, Wang, M, Zhao, Q, Wu, B. | | 登録日 | 2020-02-07 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis of ligand binding modes at the human formyl peptide receptor 2.
Nat Commun, 11, 2020
|
|
6F4V
 
 | |
6F4U
 
 | |
6V6T
 
 | |
8DKC
 
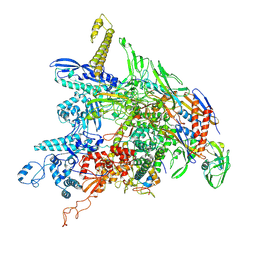 | | P. gingivalis RNA Polymerase | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Liu, B, Bu, F. | | 登録日 | 2022-07-05 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Cryo-EM Structure of Porphyromonas gingivalis RNA Polymerase.
J.Mol.Biol., 436, 2024
|
|
