7E4A
 
 | | Crystal structure of MIF bound to compound 13 | | 分子名称: | CHLORIDE ION, Flavoxate, Macrophage migration inhibitory factor, ... | | 著者 | Fa, C.P, Guo, D.Y, Yang, L. | | 登録日 | 2021-02-11 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.475 Å) | | 主引用文献 | Identification and Structure-Activity Relationships of Dietary Flavonoids as Human Macrophage Migration Inhibitory Factor (MIF) Inhibitors.
J.Agric.Food Chem., 69, 2021
|
|
7FCZ
 
 | | Crystal Structure of human RIPK1 kinase domain in complex with a novel inhibitor | | 分子名称: | N-[(3S)-7-(2-cyclopropylethynyl)-5-methyl-4-oxidanylidene-2,3-dihydro-1,5-benzoxazepin-3-yl]-5-(phenylmethyl)-4H-1,2,4-triazole-3-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | 著者 | Su, H.X, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | 登録日 | 2021-07-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Potent and Selective RIPK1 Inhibitors Targeting Dual-Pockets for the Treatment of Systemic Inflammatory Response Syndrome and Sepsis.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7FD0
 
 | | Crystal Structure of human RIPK1 kinase domain in complex with a novel inhibitor | | 分子名称: | N-[(3S)-5-methyl-7-[2-(oxan-4-yl)ethynyl]-4-oxidanylidene-2,3-dihydro-1,5-benzoxazepin-3-yl]-5-(phenylmethyl)-4H-1,2,4-triazole-3-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | 著者 | Su, H.X, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | 登録日 | 2021-07-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Potent and Selective RIPK1 Inhibitors Targeting Dual-Pockets for the Treatment of Systemic Inflammatory Response Syndrome and Sepsis.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7FAT
 
 | | Structure Determination of the RBD-NB1A7 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, nb_1A7 | | 著者 | Geng, Y, Shi, Z.Z, Li, X.Y, Wang, L, Sun, Z.C, Zhang, H.W, Chen, X.C. | | 登録日 | 2021-07-07 | | 公開日 | 2022-05-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structural basis of nanobodies neutralizing SARS-CoV-2 variants
Structure, 30, 2022
|
|
7FAU
 
 | | Structure Determination of the NB1B11-RBD Complex | | 分子名称: | NB_1B11, Spike protein S1, ZINC ION | | 著者 | Shi, Z.Z, Li, X.X, Wang, L, Sun, Z.C, Zhang, H.W, Chen, X.C, Cui, Q.Q, Qiao, H.R, Lan, Z.Y, Zhang, X. | | 登録日 | 2021-07-07 | | 公開日 | 2022-06-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Structural basis of nanobodies neutralizing SARS-CoV-2 variants.
Structure, 30, 2022
|
|
8V7Z
 
 | |
8V81
 
 | | Phosphorylated, ATP-bound, inhibitor 172-bound E1371Q human cystic fibrosis transmembrane conductance regulator | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 4-[(Z)-{(3M)-4-oxo-2-sulfanylidene-3-[3-(trifluoromethyl)phenyl]-1,3-thiazolidin-5-ylidene}methyl]benzoic acid, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Gao, X, Hwang, T. | | 登録日 | 2023-12-04 | | 公開日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Allosteric inhibition of CFTR gating by CFTRinh-172 binding in the pore
Nat Commun, 2024
|
|
4XX8
 
 | |
4P01
 
 | |
4P0H
 
 | |
4PKZ
 
 | |
4PLU
 
 | |
4QNC
 
 | | Crystal structure of a SemiSWEET in an occluded state | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, PENTADECANE, chemical transport protein | | 著者 | Yan, X, Yuyong, T, Liang, F, Perry, K. | | 登録日 | 2014-06-17 | | 公開日 | 2014-09-10 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.388 Å) | | 主引用文献 | Structures of bacterial homologues of SWEET transporters in two distinct conformations.
Nature, 515, 2014
|
|
4XX7
 
 | |
4QND
 
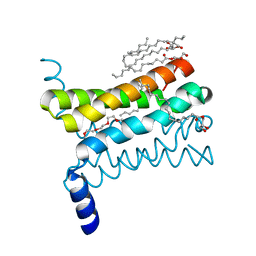 | | Crystal structure of a SemiSWEET | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Chemical transport protein, ... | | 著者 | Yan, X, Yuyong, T, Liang, F, Perry, K. | | 登録日 | 2014-06-17 | | 公開日 | 2014-09-10 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.698 Å) | | 主引用文献 | Structures of bacterial homologues of SWEET transporters in two distinct conformations.
Nature, 515, 2014
|
|
6K1T
 
 | |
6KS5
 
 | |
6L00
 
 | |
6IST
 
 | |
6LKX
 
 | | The structure of PRRSV helicase | | 分子名称: | CITRIC ACID, GLYCEROL, RNA-dependent RNA polymerase, ... | | 著者 | Shi, Y.J, Tong, X.H, Peng, G.Q. | | 登録日 | 2019-12-20 | | 公開日 | 2020-05-27 | | 最終更新日 | 2021-06-09 | | 実験手法 | X-RAY DIFFRACTION (2.998 Å) | | 主引用文献 | Structural Characterization of the Helicase nsp10 Encoded by Porcine Reproductive and Respiratory Syndrome Virus.
J.Virol., 94, 2020
|
|
5KW1
 
 | | Crystal Structure of the Two Tandem RRM Domains of PUF60 Bound to a Modified AdML Pre-mRNA 3' Splice Site Analogue | | 分子名称: | CHLORIDE ION, DNA/RNA (30-MER), Poly(U)-binding-splicing factor PUF60 | | 著者 | Crichlow, G.V, Hsiao, H.-H, Albright, R, Lolis, E.J, Braddock, D.T. | | 登録日 | 2016-07-15 | | 公開日 | 2017-08-23 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Unraveling the mechanism of recognition of the 3' splice site of the adenovirus major late promoter intron by the alternative splicing factor PUF60.
Plos One, 15, 2020
|
|
5KVY
 
 | | CRYSTAL STRUCTURE OF THE TWO TANDEM RRM DOMAINS OF PUF60 BOUND TO A PORTION OF AN ADML PRE-MRNA 3' SPLICE SITE ANALOG | | 分子名称: | CHLORIDE ION, DNA (30-MER), Poly(U)-binding-splicing factor PUF60 | | 著者 | Hsiao, H.-H, Crichlow, G.V, Albright, R.A, Murphy, J.W, Lolis, E.J, Braddock, D.T. | | 登録日 | 2016-07-15 | | 公開日 | 2017-08-23 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Unraveling the mechanism of recognition of the 3' splice site of the adenovirus major late promoter intron by the alternative splicing factor PUF60.
Plos One, 15, 2020
|
|
5KWQ
 
 | | Two Tandem RRM Domains of FBP-Interacting Repressor (FIR), also Known as PUF60 | | 分子名称: | Poly(U)-binding-splicing factor PUF60 | | 著者 | Crichlow, G.V, Yang, Y, Zhou, H, Lolis, E.J, Braddock, D.T. | | 登録日 | 2016-07-18 | | 公開日 | 2017-08-23 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Unraveling the mechanism of recognition of the 3' splice site of the adenovirus major late promoter intron by the alternative splicing factor PUF60.
Plos One, 15, 2020
|
|
5KW6
 
 | | Two Tandem RRM Domains of PUF60 Bound to an AdML Pre-mRNA 3' Splice Site Analogue with a Modified Binding-Site Nucleic Acid Base | | 分子名称: | DNA (30-MER), Poly(U)-binding-splicing factor PUF60 | | 著者 | Crichlow, G.V, Hsiao, H.-H, Albright, R, Lolis, E.J, Braddock, D.T. | | 登録日 | 2016-07-15 | | 公開日 | 2017-08-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Unraveling the mechanism of recognition of the 3' splice site of the adenovirus major late promoter intron by the alternative splicing factor PUF60.
Plos One, 15, 2020
|
|
7CAI
 
 | | SARS-CoV-2 S trimer with two RBDs in the open state and complexed with two H014 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H014 Fab, ... | | 著者 | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Wang, Y, Rao, Z, Qin, C, Wang, X. | | 登録日 | 2020-06-08 | | 公開日 | 2020-09-23 | | 最終更新日 | 2020-09-30 | | 実験手法 | ELECTRON MICROSCOPY (3.49 Å) | | 主引用文献 | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
