5DF7
 
 | | CRYSTAL STRUCTURE OF PENICILLIN-BINDING PROTEIN 3 FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH AZLOCILLIN | | 分子名称: | (2R,4S)-5,5-dimethyl-2-[(1R)-2-oxo-1-{[(2R)-2-{[(2-oxoimidazolidin-1-yl)carbonyl]amino}-2-phenylacetyl]amino}ethyl]-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, Cell division protein, ... | | 著者 | Ren, J, Nettleship, J.E, Males, A, Stuart, D.I, Owens, R.J. | | 登録日 | 2015-08-26 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structures of penicillin-binding protein 3 in complexes with azlocillin and cefoperazone in both acylated and deacylated forms.
Febs Lett., 590, 2016
|
|
5DHX
 
 | | HIV-1 Rev NTD dimers with variable crossing angles | | 分子名称: | Anti-Rev Antibody Fab single-chain variable fragment, light chain,Anti-Rev Antibody Fab single-chain variable fragment, heavy chain, ... | | 著者 | DiMattia, M.A, Watts, N.R, Wingfield, P.T, Grimes, J.M, Stuart, D.I, Steven, A.C. | | 登録日 | 2015-08-31 | | 公開日 | 2016-06-22 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | The Structure of HIV-1 Rev Filaments Suggests a Bilateral Model for Rev-RRE Assembly.
Structure, 24, 2016
|
|
5DHZ
 
 | | HIV-1 Rev NTD dimers with variable crossing angles | | 分子名称: | Anti-Rev Antibody Fab single-chain variable fragment, heavy chain, light chain, ... | | 著者 | DiMattia, M.A, Watts, N.R, Wingfield, P.T, Grimes, J.M, Stuart, D.I, Steven, A.C. | | 登録日 | 2015-08-31 | | 公開日 | 2016-06-29 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (4.3 Å) | | 主引用文献 | The Structure of HIV-1 Rev Filaments Suggests a Bilateral Model for Rev-RRE Assembly.
Structure, 24, 2016
|
|
2W0C
 
 | | X-ray structure of the entire lipid-containing bacteriophage PM2 | | 分子名称: | CALCIUM ION, MAJOR CAPSID PROTEIN P2, PROTEIN 2, ... | | 著者 | Abrescia, N.G.A, Grimes, J.M, Kivela, H.M, Assenberg, R, Sutton, G.C, Butcher, S.J, Bamford, J.K.H, Bamford, D.H, Stuart, D.I. | | 登録日 | 2008-08-13 | | 公開日 | 2008-09-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (7 Å) | | 主引用文献 | Insights Into Virus Evolution and Membrane Biogenesis from the Structure of the Marine Lipid-Containing Bacteriophage Pm2
Mol.Cell, 31, 2008
|
|
5FJ6
 
 | | Structure of the P2 polymerase inside in vitro assembled bacteriophage phi6 polymerase complex | | 分子名称: | MANGANESE (II) ION, RNA-DIRECTED RNA POLYMERASE | | 著者 | Ilca, S, Kotecha, A, Sun, X, Poranen, M.P, Stuart, D.I, Huiskonen, J.T. | | 登録日 | 2015-10-06 | | 公開日 | 2015-11-04 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7.9 Å) | | 主引用文献 | Localized Reconstruction of Subunits from Electron Cryomicroscopy Images of Macromolecular Complexes.
Nat.Commun., 6, 2015
|
|
8CI3
 
 | | Structure of bovine CD46 ectodomain (SCR 1-2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | 著者 | Aitkenhead, H, David I Stuart, D.I, El Omari, K. | | 登録日 | 2023-02-08 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Structure of Bovine CD46 Ectodomain.
Viruses, 15, 2023
|
|
5FJ7
 
 | | Structure of the P2 polymerase inside in vitro assembled bacteriophage phi6 polymerase complex, with P1 included | | 分子名称: | MAJOR INNER PROTEIN P1, MANGANESE (II) ION, RNA-DIRECTED RNA POLYMERASE | | 著者 | Ilca, S, Kotecha, A, Sun, X, Poranen, M.P, Stuart, D.I, Huiskonen, J.T. | | 登録日 | 2015-10-06 | | 公開日 | 2015-11-04 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7.9 Å) | | 主引用文献 | Localized Reconstruction of Subunits from Electron Cryomicroscopy Images of Macromolecular Complexes.
Nat.Commun., 6, 2015
|
|
5FJ5
 
 | | Structure of the in vitro assembled bacteriophage phi6 polymerase complex | | 分子名称: | MAJOR INNER PROTEIN P1 | | 著者 | Ilca, S, Kotecha, A, Sun, X, Poranen, M.P, Stuart, D.I, Huiskonen, J.T. | | 登録日 | 2015-10-06 | | 公開日 | 2015-11-04 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Localized Reconstruction of Subunits from Electron Cryomicroscopy Images of Macromolecular Complexes.
Nat.Commun., 6, 2015
|
|
8CII
 
 | | Delta-RBD complex with BA.2-07 fab, SARS1-34 fab and C1 nanobody | | 分子名称: | BA.2-07 fab Heavy Chain, BA.2-07 fab Light Chain, C1 nanobody, ... | | 著者 | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I, Fry, E.E. | | 登録日 | 2023-02-09 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Potent cross-reactive mAbs from BA.4/5 breakthrough infection
To Be Published
|
|
8CIN
 
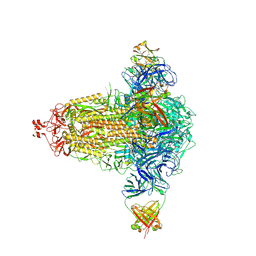 | | BA.4/5-5 FAB IN COMPLEX WITH SARS-COV-2 BA.4 SPIKE GLYCOPROTEIN | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA.4/5-5 fab HEAVY CHAIN, ... | | 著者 | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I, Fry, E.E. | | 登録日 | 2023-02-10 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | The SARS-CoV-2 neutralizing antibody response to SD1 and its evasion by BA.2.86.
Nat Commun, 15, 2024
|
|
8BP8
 
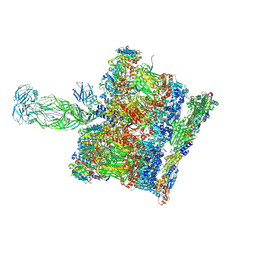 | | SPA of Trypsin untreated Rotavirus TLP spike | | 分子名称: | CALCIUM ION, Inner capsid protein VP2, Intermediate capsid protein VP6, ... | | 著者 | Shah, P.N.M, Stuart, D.I. | | 登録日 | 2022-11-16 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Characterization of the rotavirus assembly pathway in situ using cryoelectron tomography.
Cell Host Microbe, 31, 2023
|
|
8COA
 
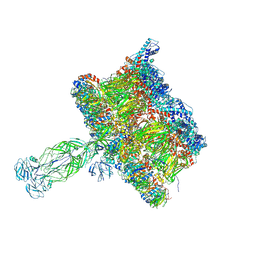 | |
8CO6
 
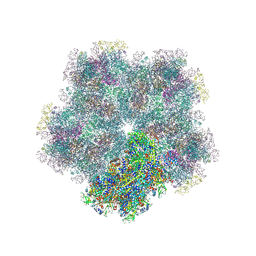 | |
8CIM
 
 | | BA.2-07 FAB IN COMPLEX WITH SARS-COV-2 BA.2.12.1 SPIKE GLYCOPROTEIN | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA.2-07 FAB HEAVY CHAIN, ... | | 著者 | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I, Fry, E.E. | | 登録日 | 2023-02-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Isolation of a pair of potent broadly neutralizing mAb binding to RBD and SD1 domains of SARS-CoV-2
Res Sq, 2023
|
|
1VCA
 
 | | CRYSTAL STRUCTURE OF AN INTEGRIN-BINDING FRAGMENT OF VASCULAR CELL ADHESION MOLECULE-1 AT 1.8 ANGSTROMS RESOLUTION | | 分子名称: | HUMAN VASCULAR CELL ADHESION MOLECULE-1 | | 著者 | Jones, E.Y, Harlos, K, Bottomley, M.J, Robinson, R.C, Driscoll, P.C, Edwards, R.M, Clements, J.M, Dudgeon, T.J, Stuart, D.I. | | 登録日 | 1995-03-21 | | 公開日 | 1995-09-15 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of an integrin-binding fragment of vascular cell adhesion molecule-1 at 1.8 A resolution.
Nature, 373, 1995
|
|
4X35
 
 | | A micro-patterned silicon chip as sample holder for macromolecular crystallography experiments with minimal background scattering | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Roedig, P, Vartiainen, I, Duman, R, Panneerselvam, S, Stuebe, N, Lorbeer, O, Warmer, M, Sutton, G, Stuart, D.I, Weckert, E, David, C, Wagner, A, Meents, A. | | 登録日 | 2014-11-27 | | 公開日 | 2015-06-10 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A micro-patterned silicon chip as sample holder for macromolecular crystallography experiments with minimal background scattering.
Sci Rep, 5, 2015
|
|
6G9B
 
 | | Crystal structure of Ebolavirus glycoprotein in complex with imipramine | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(5H-DIBENZO[B,F]AZEPIN-5-YL)-N,N-DIMETHYLPROPAN-1-AMINE, ... | | 著者 | Zhao, Y, Ren, J, Fry, E.E, Xiao, J, Townsend, A.R, Stuart, D.I. | | 登録日 | 2018-04-10 | | 公開日 | 2018-05-23 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Structures of Ebola Virus Glycoprotein Complexes with Tricyclic Antidepressant and Antipsychotic Drugs.
J. Med. Chem., 61, 2018
|
|
6G95
 
 | | Crystal structure of Ebolavirus glycoprotein in complex with thioridazine | | 分子名称: | 10-{2-[(2R)-1-methylpiperidin-2-yl]ethyl}-2-(methylsulfanyl)-10H-phenothiazine, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | 著者 | Zhao, Y, Ren, J, Fry, E.E, Xiao, J, Townsend, A.R, Stuart, D.I. | | 登録日 | 2018-04-10 | | 公開日 | 2018-05-23 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Structures of Ebola Virus Glycoprotein Complexes with Tricyclic Antidepressant and Antipsychotic Drugs.
J. Med. Chem., 61, 2018
|
|
6G9I
 
 | | Crystal structure of Ebolavirus glycoprotein in complex with clomipramine | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(3-CHLORO-5H-DIBENZO[B,F]AZEPIN-5-YL)-N,N-DIMETHYLPROPAN-1-AMINE, Envelope glycoprotein,Envelope glycoprotein, ... | | 著者 | Zhao, Y, Ren, J, Fry, E.E, Xiao, J, Townsend, A.R, Stuart, D.I. | | 登録日 | 2018-04-10 | | 公開日 | 2018-05-23 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Structures of Ebola Virus Glycoprotein Complexes with Tricyclic Antidepressant and Antipsychotic Drugs.
J. Med. Chem., 61, 2018
|
|
6YB7
 
 | | SARS-CoV-2 main protease with unliganded active site (2019-nCoV, coronavirus disease 2019, COVID-19). | | 分子名称: | 3C-like proteinase, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE | | 著者 | Owen, C.D, Lukacik, P, Strain-Damerell, C.M, Douangamath, A, Powell, A.J, Fearon, D, Brandao-Neto, J, Crawshaw, A.D, Aragao, D, Williams, M, Flaig, R, Hall, D.R, McAuley, K.E, Mazzorana, M, Stuart, D.I, von Delft, F, Walsh, M.A. | | 登録日 | 2020-03-16 | | 公開日 | 2020-03-25 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | COVID-19 main protease with unliganded active site
To Be Published
|
|
6Y84
 
 | | SARS-CoV-2 main protease with unliganded active site (2019-nCoV, coronavirus disease 2019, COVID-19) | | 分子名称: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | 著者 | Owen, C.D, Lukacik, P, Strain-Damerell, C.M, Douangamath, A, Powell, A.J, Fearon, D, Brandao-Neto, J, Crawshaw, A.D, Aragao, D, Williams, M, Flaig, R, Hall, D.R, McAuley, K.E, Mazzorana, M, Stuart, D.I, von Delft, F, Walsh, M.A. | | 登録日 | 2020-03-03 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | COVID-19 main protease with unliganded active site
To Be Published
|
|
6H3B
 
 | |
4X3B
 
 | | A micro-patterned silicon chip as sample holder for macromolecular crystallography experiments with minimal background scattering | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Roedig, P, Vartiainen, I, Duman, R, Panneerselvam, S, Stuebe, N, Lorbeer, O, Warmer, M, Sutton, G, Stuart, D.I, Weckert, E, David, C, Wagner, A, Meents, A. | | 登録日 | 2014-11-28 | | 公開日 | 2015-06-10 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A micro-patterned silicon chip as sample holder for macromolecular crystallography experiments with minimal background scattering.
Sci Rep, 5, 2015
|
|
6YM0
 
 | | Crystal structure of the SARS-CoV-2 receptor binding domain in complex with CR3022 Fab (crystal form 1) | | 分子名称: | Spike glycoprotein, heavy chain, light chain | | 著者 | Huo, J, Zhao, Y, Ren, J, Zhou, D, Ginn, H.M, Fry, E.E, Owens, R, Stuart, D.I. | | 登録日 | 2020-04-07 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (4.36 Å) | | 主引用文献 | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
6YOR
 
 | | Structure of the SARS-CoV-2 spike S1 protein in complex with CR3022 Fab | | 分子名称: | IgG H chain, IgG L chain, Spike glycoprotein | | 著者 | Huo, J, Zhao, Y, Ren, J, Zhou, D, Duyvesteyn, H.M.E, Carrique, L, Malinauskas, T, Ruza, R.R, Shah, P.N.M, Fry, E.E, Owens, R, Stuart, D.I. | | 登録日 | 2020-04-15 | | 公開日 | 2020-04-29 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
